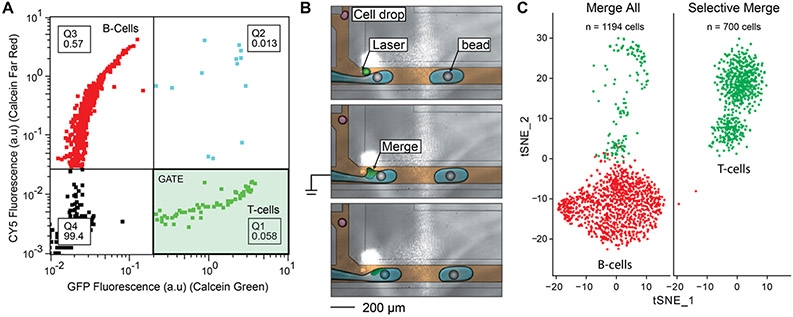
Figure 3.
Single-cell RNA seq of an immune subpopulation using the fluorescence-activated droplet merger. (A) Cytometer plot of droplet fluorescence. Four quadrants (Q1–Q4) are defined, and the percentage of drops in each quadrant is displayed. Q1 contains calcein green positive drops, Q2 contains calcein green and CellTrace Far red positive drops, Q3 contains CellTrace Far red positive drops, and Q4 contains empty drops. The Q1 gate (shaded green) is used to selectively merge T-cells. (B) Cells are merged with a stream of barcoded beads for mRNA capture and reverse transcription. Only merged cells are co-encapsulated with beads and reagents for cDNA synthesis. The stream immediately forms droplets at a T-junction downstream of the merging event. The cell drops are colored red (negative) and green (positive, merge), PCR reagents are colored blue, barcoded beads are colored gray, and oil is colored brown. (C) Single-cell RNA-seq and t-SNE clustering confirms that the desired subpopulation is targeted for sequencing. Merging all of the cells with barcoded beads performs scRNAseq on both B-cell and T-cells, while merging onlyT-cells targets that population.