Abstract
Objective:
To develop and evaluate novel word embeddings (WEs) specific to ophthalmology, using text corpora from published literature and electronic health records (EHR).
Materials and Methods:
We trained ophthalmology-specific WEs using 121,740 PubMed abstracts and 89,282 EHR notes using word2vec continuous bag-of-words architecture. PubMed and EHR WEs were compared to general domain GloVe WEs and general biomedical domain BioWordVec embeddings using a novel ophthalmology-domain-specific 200-question analogy test and prediction of prognosis in 5547 low vision patients using EHR notes as inputs to a deep learning model.
Results:
We found that many words representing important ophthalmic concepts in the EHR were missing from the general domain GloVe vocabulary, but covered in the ophthalmology abstract corpus. On ophthalmology analogy testing, PubMed WEs scored 95.0%, outperforming EHR (86.0%) and GloVe (91.0 %) but less than BioWordVec (99.5%). On predicting low vision prognosis, PubMed and EHR WEs resulted in similar AUROC (0.830; 0.826), outperforming GloVe (0.778) and BioWordVec (0.784).
Conclusion:
We found that using ophthalmology domain-specific WEs improved performance in ophthalmology-related clinical prediction compared to general WEs. Deep learning models using clinical notes as inputs can predict the prognosis of visually impaired patients. This work provides a framework to improve predictive models using domain-specific WEs.
Keywords: ophthalmology, natural language processing, deep learning, vision, low, informatics
1. INTRODUCTION
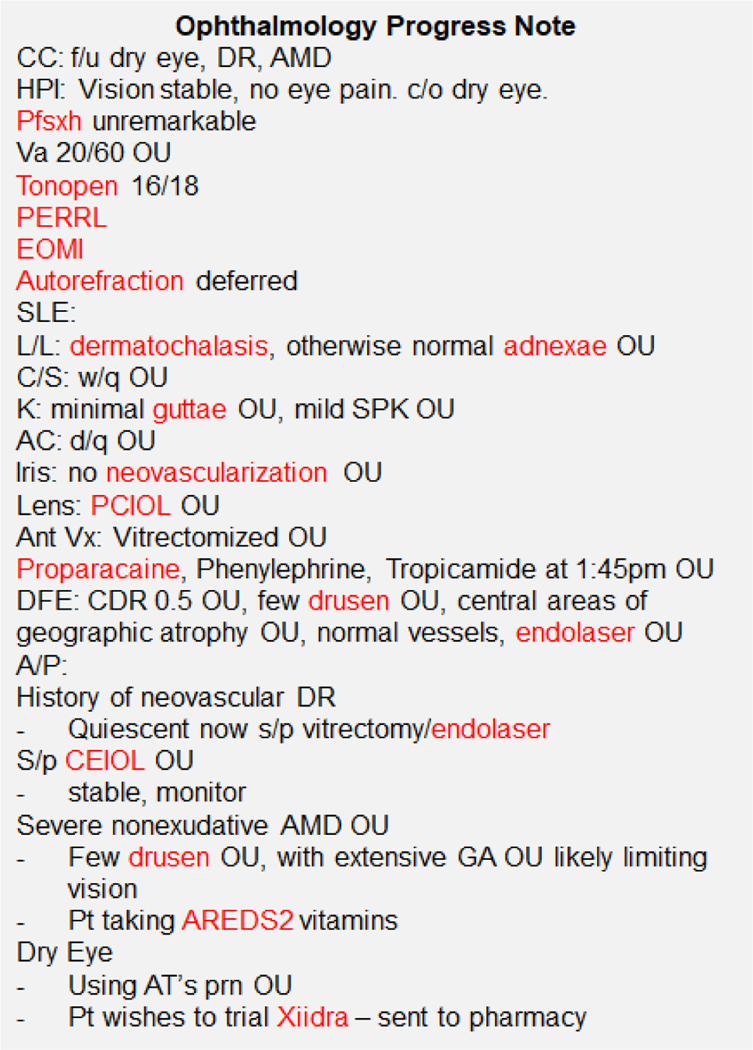
With the widespread adoption of electronic health records (EHR), informatics techniques are increasingly used to mine this rich source of information to build prediction algorithms, including in the field of ophthalmology.[1–3] However, much of the clinical information is captured in unstructured free text using highly specialized domain-specific language and abbreviations (Figure 1).[4,5] In ophthalmology, this includes crucial information on eye examination findings which are important indicators of disease severity and prognosis, which are difficult to incorporate into prediction models as free text.
Figure 1. Example Ophthalmology Progress Note.
Legend: There are many specialized and domain-specific terms and abbreviations, such that even physicians in other specialties would have difficulty understanding ophthalmology progress notes. Words highlighted in red are those which occur very commonly in ophthalmology but are not in the vocabulary of general-domain word embeddings (GloVe).
In particular, in ophthalmology there is a need to develop algorithms that can predict the visual prognosis of patients with visual impairment, in order to better enable the targeting of important resources, such as multidisciplinary low vision rehabilitation services,[6] to those patients most likely to benefit. Almost 3 million adults in the United States are estimated to have irreversible low vision and would benefit from such rehabilitation services to improve their quality of life and daily functioning,[6] but the referral rate is extraordinarily low, leaving almost 90% of patients who may benefit without access to or awareness of these services[7], which may be due to a variety of reasons including limited time during clinic visits to introduce these services, or optimistic assumptions that vision will soon improve with therapy. Using information from EHR to identify vision rehabilitation candidates in an automated manner could better facilitate timely referrals to improve access to these important services. Such predictive algorithms could detect particular findings or diagnoses documented in clinical free text notes which are known not to be reversible, such as retinal atrophy associated with macular degeneration, or predict for particular patients that they would not achieve vastly improved vision with treatment.
The use of neural word embeddings is an approach to incorporating biomedical text into prediction models, as word embeddings enable computation over free-text by representing word meaning as dense numerical vectors. General word embeddings enabled breakthroughs in performance on named entity recognition, sentence classification, relation extraction, and other general natural language processing tasks,[8] and general domain GloVe (Global Vectors for Word Representation), pre-trained on large corpora of general content such as Common Crawl (general internet pages)[9–12], are publicly available. However, use of general word embeddings in the biomedical domain may be hampered by the concern that many biomedical terms would appear so infrequently in conventional corpora that pre-trained word embeddings for those terms may not be meaningful. In addition, out-of-vocabulary terms which do not appear at all in the corpus vocabulary do not have a meaningful word embedding. Word vectors trained on large corpora of general biomedical text have also been developed.[13,14] However, application to even more subspecialized biomedical domains such as ophthalmology, with its own very rich set of abbreviations and terminology,[4,5] may pose an especial challenge due to differences in vocabulary and their usage.
In ophthalmology and in other biomedical domains, a major challenge to the use of EHR to develop predictive algorithms is the inability to incorporate the wealth of information sequestered within the clinical free-text, and the use of domain-specific neural word embeddings may provide one solution. The objective of this study was to train and evaluate word vectors specific to the ophthalmology domain using corpora from published ophthalmology literature and from EHRs, comparing them to embeddings pre-trained on more general corpora. We include evaluation of ophthalmology word embeddings on intrinsic tasks, including on a novel set of ophthalmology domain-specific analogies developed for this purpose, as well as evaluation on an extrinsic prediction task to predict the visual prognosis of visually impaired patients using clinical free-text notes from the EHR. We hypothesized that using ophthalmology domain-specific word embeddings would result in better performance on ophthalmology-related tasks than using more general word embeddings. The work we present provides a framework for training and evaluating domain-specific word embeddings that can be generalized to many domains across medicine and applied to a variety of clinical prediction tasks.
2. METHODS
2.1. Training Ophthalmology Domain-Specific Word Embeddings:
2.1.1. PubMed Ophthalmology Embedding Corpus:
We extracted all English-language abstracts indexed in PubMed from 2009–2019 belonging to the MeSH categories of “Eye Diseases,” “Ocular Physiological Phenomena”, “Ophthalmology”, “Ophthalmologic Surgical Procedures,” or their subcategories. Animal studies were excluded. Abstracts shorter than 50 characters, with an associated title shorter than 3 characters, or with no listed authors or journal were excluded. In total there were 121,740 ophthalmology abstracts included.
2.1.2. Electronic Health Records Ophthalmology Embeddings Corpus:
We identified all ophthalmology clinical notes from Stanford STARR[15,16] of length > 50 characters. As clinical notes are often copied forward from visit to visit for each patient, resulting in highly repetitive text, we randomly sample one clinical note for each unique patient, resulting in a corpus of 89,282 ophthalmology EHR notes. This study received approval from the Institutional Review Board (IRB) of the participating institution.
2.1.3. Corpora Processing and Model Training:
The PubMed and EHR corpora were pre-processed in identical fashion. All words were lowercase and tokenized. Common stopwords were removed (‘a’, ‘all’, ‘also’, ‘an’, ‘and’, ‘are’, ‘as’, ‘at’, ‘be’, ‘been’, ‘by’, ‘for’, ‘from’, ‘had’, ‘has’, ‘have’, ‘in’, ‘is’, ‘it’, ‘may’, ‘of’, ‘on’, ‘or’, ‘our’, ‘than’, ‘that’, ‘the’, ‘there’, ‘these’, ‘this’, ‘to’, ‘was’, ‘we’, ‘were’, ‘which’, ‘who’, ‘with’). In all there were 55,937 tokens in the PubMed corpus and 41,630 tokens in the EHR corpus which appeared with frequency ≥ 5 in their respective corpora. Embeddings were trained with the established word2vec neural network architecture[11] for the continuous-bag-of-words task which predicts a target word given a context window. The embedding dimension was set to 300 to facilitate comparison to baseline GloVe vectors (see below). The word window size was set to 5. Models were trained using Tensorflow (version 2.1.0).
2.1.4. Baseline Embedding Comparisons:
We used as our baseline comparisons uncased 300-dimensional GloVe vectors, covering 42 billion tokens trained on the Common Crawl,[9,10] and the 200-dimensional BioWordVec vectors previously trained on PubMed biomedical literature and MMIC-III containing EHR data from inpatient ICU hospitalization notes.[13,14]
2.2. Evaluation of Word Embeddings:
Word embeddings can be evaluated on an “intrinsic” basis, so-called because “intrinsic” evaluation only relies upon evaluation of the structure of the word embeddings themselves, as well as on an “extrinsic” basis, which evaluates performance upon downstream applications of the word embeddings to specific external tasks.[17,18]
2.2.1. Intrinsic Evaluation
2.2.1.1. Examining Vocabulary and Principal Component Visualization of Example Clusters
We examined vocabulary that was common in the EHR corpus but that was not in PubMed, BioWordVec, or GloVe corpus, to identify potential gaps in coverage for important clinical concepts. After training of the ophthalmology word embeddings, we also qualitatively examined whether similar concepts were clustered appropriately in ophthalmology embedding space. We chose three seed terms, “poag”, “orbit”, and “guttae”, and determined their 9 nearest neighbors in the corpus in the full embedding space. These three clusters of 10 embeddings each were then visualized in a 2-dimensional projection onto the first and second principal components of embedding space. These seed terms were chosen as they come from different subspecialties of ophthalmology; “poag” is an abbreviation for primary open angle glaucoma and would be most relevant to the glaucoma subspecialty, “orbit” would be most commonly encountered in the context of oculoplastics subspecialty notes, and “guttae” is a finding of the cornea that would be most commonly encountered in the cornea subspecialty.
2.2.1.2. Ophthalmology Domain-Specific Analogies
General analogies developed to evaluate word vectors cannot be easily extended for use in subdomains such as ophthalmology as they do not adequately or accurately test ophthalmology-related concepts. For example, only a small fraction of general biomedical domain analogies are relevant to ophthalmology. Thus, we developed novel ophthalmology domain-specific 200-question analogy test to perform formal intrinsic evaluation of our word embeddings. Ophthalmology-related word pairs with analogous semantic relations were matched by a board-certified ophthalmologist, identified from words that were common to all three sets of embeddings. These word pairs were matched into analogies, for example, an analogy constructed as word1:word2::word3:word4 might have an example of conjunctiva:conjunctival::eyelid:palpebral. For each correct analogy, a random wrong word was chosen from the analogy vocabulary to serve as a wrong answer choice compared to word4, the correct analogy completion choice. All analogies as well as the random wrong word choice were manually reviewed for semantic validity. Final analogies are publicly available.[19] For the analogy test, the cosine similarity between (word2-word1+word3, word4) and (word2-word1+word3, wrongword) was calculated in the PubMed, EHR, BioWordVec, and GloVe embedding spaces, and the closer word choice was determined to be the “answer” for that embedding for that analogy question. Accuracy on the analogy test was calculated as wrong answers / total number of questions (N=200).
2.2.2. Extrinsic Evaluation: Predicting Low Vision Prognosis
To evaluate the performance of our domain-specific embeddings on an extrinsic task, we mapped words from EHR free-text clinical notes to neural word embeddings to use as input features for a deep learning model to predict the visual prognosis in a cohort of patients with visual impairment.
2.2.2.1. Cohort Definition
We identified from the Stanford Clinical Data Warehouse[15] all documented visual acuity measurements (N=553,184) belonging to N=88,692 unique adult patients from 2009–2018.[16] Visual acuity measurements were captured from semi-structured fields, including distance, near, with refraction, with or without habitual glasses or contacts for either eye, using a combination of rule-based algorithms based on regular expressions[16]. Low vision on a particular encounter date was defined as visual acuity worse than 20/40 on all visual acuity measurements documented for that encounter. If only the visual acuity of one eye was measured for that encounter date, as may be common in a postoperative visit focused on one eye, then the most recent previous visual acuity for the contralateral eye was used to forward fill the missing value for that encounter. In total there were N=13,847 patients with at least one documented encounter with low vision. The first date of low vision was determined for each patient (hereafter referred to as the index date). We included patients with follow-up for at least one year from the index date, defined as ≥ one visit with documented visual acuity measurement ≥ 365 days from the index date (N=5612). For these patients, we extracted all ophthalmology free-text clinical notes on or prior to the index date (N=5547 patients with available notes).
2.2.2.2. Modeling approach
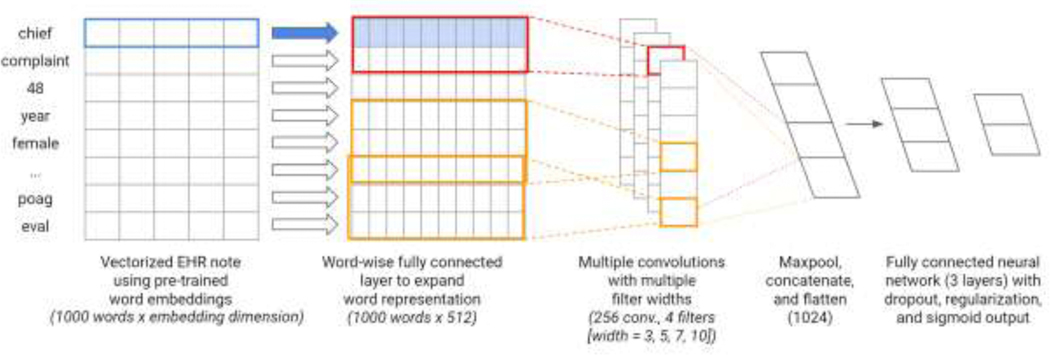
The prediction task was to determine whether low vision patients would still have poor visual acuity (<20/40) after one year or follow-up, indicating a poor visual prognosis that may benefit from referral to low vision rehabilitation services aimed at improving quality of life and activities of daily living of visually impaired patients by delivering interventions to maximize the function of the remaining vision. Overall, 40.7% (N=2,258) of patients did not improve to 20/40 or better within one year. The model architecture is depicted in Figure 2. The overall architecture is based on a previously published TextCNN architecture,[20] which utilizes multiple convolutions with multiple filter widths to convolve over word sequences of different lengths, thus capturing some information regarding the context in which words are used. We used as inputs to the models clinical free text notes on or prior to the date of low vision, as these would be the same information available to clinicians presented with a low vision patient. Because the amount of historical clinical documentation varied between patients, we arranged all notes in backwards chronological order on the premise that more recent notes would hold more relevant information for prognosis prediction. The most recent 1000 words of clinical documentation were mapped to word embeddings (either EHR, PubMed, BioWordVec, or GloVe) and used as inputs to the deep learning model. This length of input text was arbitrarily chosen to be close to the median length of documentation for each patient (923 words). Words that were missing from embedding vocabularies were mapped to a generic token for unknown words. The embedding layer was followed by a fully connected layer to expand the representation matrix to shape 1000 × 512 and a dropout layer (rate = 0.5). After following hyperparameter tuning procedures as described by Zhang et al [21], we arrived at a model architecture which used 4 convolutions of region size 3, 5, 7, and 10 with 256 filters each, followed by a max pooling layer. The resulting vectors are concatenated and flattened and followed by a dropout layer (rate = 0.5), a fully connected layer with L2 regularization (alpha = 0.01), a subsequent dropout layer (rate = 0.5), an additional fully connected layer with L2 regularization (alpha = 0.01) and a final sigmoid output for the prediction. We randomly selected 6% of the training data as the validation set for early stopping.
Figure 2. Visual Prognosis Prediction Task Model Architecture.
Legend: Words from clinical progress notes were mapped to word embeddings (either EHR, PubMed, BioWordVec or GloVe) and used as inputs to a deep learning model. Multiple convolutions with multiple filter widths were passed over the word representation matrix followed by max pooling, concatenation, and flattening operations. Subsequent fully connected layers included dropout, regularization, and a final sigmoid output to predict the binary outcome of whether patients would have persistent poor visual acuity after one year of follow-up.
2.2.2.3. Model Evaluation
The cohort was split into validation and test sets of 300 patients each, with the remainder reserved for training. Final performance metrics for all models included the standard measures of prediction accuracy, sensitivity (recall), specificity, positive predictive value (precision), negative predictive value, and F1 score (the harmonic mean of precision and recall). We also calculated the area under the receiver operating curve (AUROC) and area under the precision-recall curve (AUPRC). In addition, notes from a random subset of the test set of patients (N=102) were evaluated by a board-certified ophthalmologist (SYW) to provide a human-level performance baseline for evaluation metrics.
2.2.2.3.1. Code Availability
All code used to train and evaluate ophthalmology word embeddings is available in a public code repository.[19] PubMed ophthalmology word embeddings are also available for download. Due to the potential sensitive patient health information contained in words in the EHR, our EHR word embeddings are not included. The full set of ophthalmology domain-specific analogies is also included for reuse. Finally, code for training the deep learning algorithm to predict visual prognosis is also included in the repository.
3. RESULTS
3.1. Word Embedding Vocabulary
There were 41,630 unique words and 55,937 unique words that appeared with frequency ≥ 5 in the EHR and PubMed corpora, respectively. A total of 4370 unique words appeared in the EHR corpus at least 5 times which did not appear in GloVe, while a total of 20,894 unique words appeared in the EHR corpus a minimum of 5 times which did not appear in the PubMed embeddings at least 5 times. A total of 3354 unique words that appeared in the EHR corpus at least 5 times were not in BioWordVec. The most common of these words along with their frequency of appearance is summarized in Table 1.
Table 1:
Vocabulary Comparison between Word Embeddings
| Words in EHR but not in GloVe | Words in EHR but not in PubMed Embeddings | Words in EHR but not in BioWordVec | |||
|---|---|---|---|---|---|
| Word | Frequency | Word | Frequency | Word | Frequency |
| tonopen | 52650 | hx | 71719 | {redacted, physician name} | 12751 |
| autorefraction | 20272 | psh | 55803 | {redacted, physician name} | 9805 |
| adnexae | 17195 | disp | 53091 | {redacted, zip code} | 6972 |
| perrl | 10633 | rfl | 50736 | pfshx | 5804 |
| pciol | 7969 | reconciliation | 33314 | {redacted, zip code} | 4392 |
| pseudophakia | 7186 | dob | 31171 | {redacted, physician name} | 4217 |
| pfshx | 5804 | dear | 29437 | {redacted, physician name} | 3156 |
| dermatochalasis | 5652 | 6995 | 28153 | cannot | 2794 |
| proparacaine | 4876 | thank | 27984 | cuie2 | 2204 |
| lissamine | 3683 | accomodation | 26110 | eoph453 | 2204 |
| eomi | 3043 | {redacted, zip code} | 21635 | basename | 2035 |
| {redacted, physician name} | 3156 | sincerely | 19479 | {redacted, physician name} | 1987 |
| cclist | 2911 | csn | 19446 | {redacted, zip code} | 1794 |
| hypertropia | 2646 | dist | 17459 | {redacted, zip code} | 1544 |
| orthophoric | 2567 | meds | 13694 | {redacted, zip code} | 1200 |
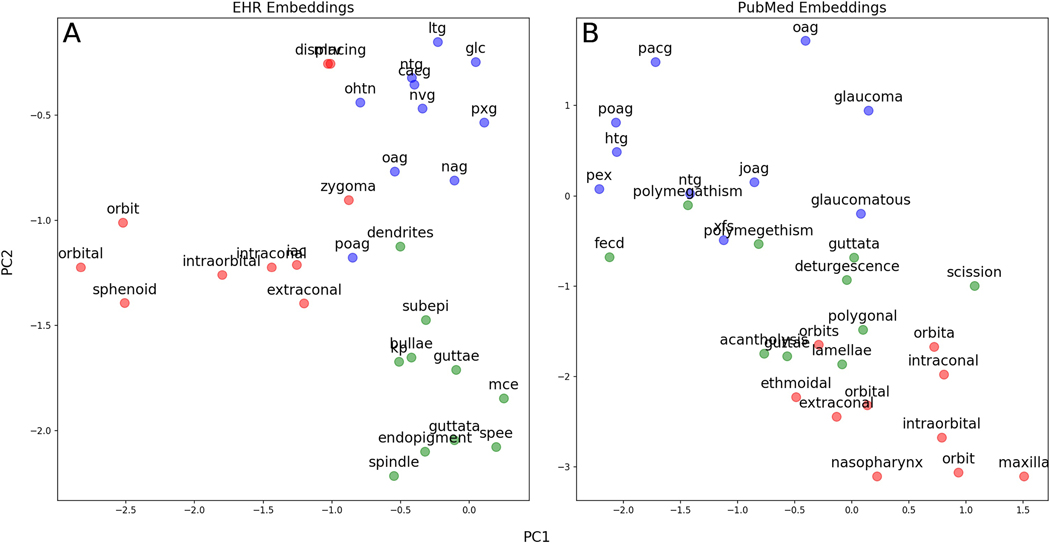
We used principal component analysis to project onto two dimensions the word embeddings for the 10 closest words clustered around three separate ophthalmology concepts (Figure 3). For both EHR and PubMed embeddings, words embeddings close to poag included a variety of abbreviations for different forms of glaucoma; those close to orbit included anatomical structures near the orbit. In EHR embeddings, words close to guttae included other findings common in the cornea; in PubMed embeddings, words close to guttae included words often used to describe diseased corneal endothelial cells (“polymegathism”, “polygonal”) and Fuch’s endothelial corneal dystrophy (“fecd”), which all result in the finding of guttae.
Figure 3. Clusters of Word Embeddings Projected onto the First Two Principal Components.
Legend: The 10 closest words in embedding space to poag (blue), orbit (red), and guttae (green) are projected into the first two principal components of the embedding space for A) EHR embeddings and B) PubMed embeddings. In both panels, terms from these different ophthalmology subspecialties cluster appropriately in different areas of embedding space. Words are similar between the two sets of embeddings, although PubMed words closest to guttae contain more words often used to describe diseased corneal endothelial cells in the scientific literature (polymegathism, polygonal) rather than in clinical use.
3.2. Novel Ophthalmology Domain-Specific Analogies
We created a novel set of 200 ophthalmology-related analogies to evaluate the intrinsic performance of these word embeddings on an ophthalmology-specific task. Examples of analogies are shown in Table 3, with the full set publicly available.[19] On analogy testing, PubMed WEs scored 95.0% accuracy, outperforming EHR (86.0%) and GloVe (91.0 %). BioWordVec WEs scored 99.5% accuracy, outperforming all other embeddings. An example analogy with correct and incorrect answer choices is depicted in embedding spaces in Figure 4.
Table 3.
Examples of Ophthalmology Domain-Specific Analogies
| Relationship | Word 1 | Word 2 | Word 3 | Word 4 |
|---|---|---|---|---|
| Sister drugs within the same class | dorzolamide | brinzolamide | bromfenac | ketorolac |
| Anatomical locations and their adjectives | limbus | limbal | canthus | canthal |
| Diseases and their affected anatomy | ocp | conjunctiva | pseudoexfoliation | lens |
| Anatomical locations and inflammatory conditions at that location |
uvea | uveitis | choroid | choroiditis |
| Antonyms | photopic | scotopic | light | dark |
| Laterality | left | right | os | od |
| Drugs and the disease they treat | brinzolamide | glaucoma | bevacizumab | amd |
| Anatomical locations and procedures performed at that location |
sclera | sclerotomy | iris | iridotomy |
Figure 4. Example Ophthalmology-Specific Analogy Test Question in Embedding Space.
Legend: An example analogy question (ocp:conjunctiva::pseudoexfoliation::???) is depicted in a two-dimensional principal component projection of word embedding space. The correct word for analogy completion is lens, whereas the randomly chosen incorrect word is neuritis. Cosine similarity is calculated between (pseudoexfoliation - ocp + conjunctiva) and either lens or neuritis and the closer word in embedding space is chosen as the answer. In this example, (A) PubMed, (B) EHR, and (C) BioWordVec word embeddings identify the correct answer while (D) GloVe chooses the wrong answer.
3.3. Extrinsic Evaluation: Predicting Low Vision Prognosis in Electronic Health Records Using Word Embeddings
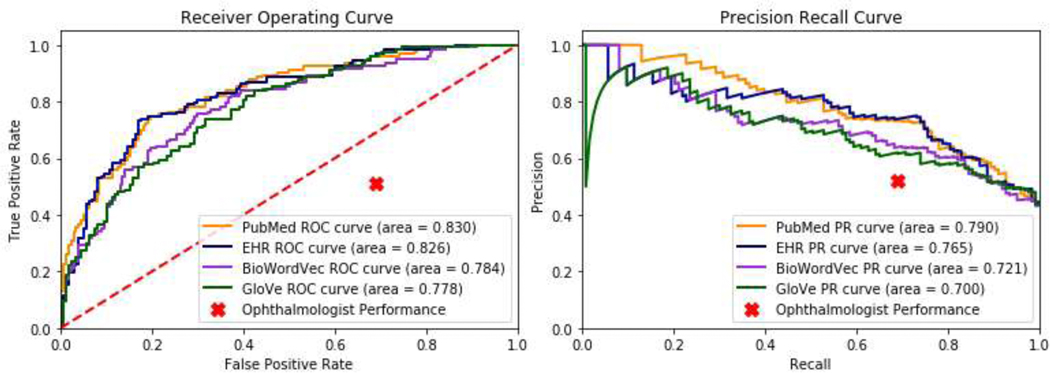
To compare the results of using different types of word embeddings on an extrinsic evaluation task, we developed deep learning models to predict visual prognosis using clinical progress notes from EHR. Words from free text clinical progress notes for a cohort of low vision patients were mapped to either PubMed, EHR, BioWordVec, or GloVe word embeddings and used as inputs to a deep learning model to predict whether patients would still have poor vision after 1 year of follow-up. Using PubMed and EHR WEs resulted in similar AUROC (0.830; 0.826), outperforming GloVe (0.778) and BioWordVec (0.784). Receiver operating and precision recall curves are depicted in Figure 5. Additional performance metrics at the classification probability threshold of 50% are shown in Table 4.
Figure 5: Receiver Operating and Precision-Recall Curves for Prediction of Low Vision Prognosis.
Legend: (A) Receiver operating and (B) precision-recall curves are shown for deep learning models predicting low vision prognosis which use as inputs free text clinical notes mapped to either more general GloVe or BioWordVec word embeddings, or custom ophthalmology-domain embeddings trained on either PubMed ophthalmology abstracts or ophthalmology clinical free text notes from the electronic health records (EHR). Ophthalmologist prediction performance is shown as a single point.
Table 4:
Performance Metrics for Prediction of Visual Prognosis Using Various Word Embeddings
| Pubmed | EHR | BioWordVec | GloVe | Ophthalmologist | |
|---|---|---|---|---|---|
| F1 | 0.73 | 0.72 | 0.63 | 0.65 | 0.59 |
| Sensitivity (Recall) | 0.76 | 0.77 | 0.59 | 0.74 | 0.69 |
| Specificity | 0.78 | 0.76 | 0.82 | 0.63 | 0.49 |
| PPV (Precision) | 0.70 | 0.68 | 0.69 | 0.58 | 0.52 |
| NPV | 0.82 | 0.82 | 0.74 | 0.78 | 0.67 |
| Accuracy | 0.77 | 0.76 | 0.72 | 0.68 | 0.58 |
4. DISCUSSION
Predicting the prognosis of visual impairment is a challenge in ophthalmology and the lack of ophthalmology-specific embeddings may contribute to this challenge. In this study, we trained ophthalmology-specific embeddings using EHR and PubMed text corpora. We found that EHR and PubMed embeddings perform similarly, yet better than more general word embeddings on an extrinsic evaluation with an ophthalmology-specific clinical predictive task using EHR free-text progress notes. Using only clinical progress notes as inputs, deep learning models using ophthalmology-specific embeddings were able to perform with relatively good AUROC (>0.80) to predict the prognosis of visually impaired patients in a held-out test set, indicating that this may be a promising approach to using ophthalmology clinical notes for clinical predictive tasks.
For formal intrinsic comparison of ophthalmology domain-specific embeddings to general embeddings, we developed a novel ophthalmology domain-specific analogy test.[19] An advantage of using analogies to evaluate embeddings is that evaluation is easy to perform, not computationally intensive, and does not require the curation and labeling of a dataset for a specific downstream clinical task. Because the corpus of all biomedical terms is vast, and the proportion of ophthalmology-related words is relatively small, previous approaches in developing general biomedical domain analogies[18,22] using word pairs from general biomedical ontologies was not an appropriate method of evaluating embeddings created specifically for ophthalmology-related tasks. In addition, programmatically matching word pairs to create analogies more than occasionally created analogies which were semantically invalid. For example, in one prior set of biomedical analogies, BMASS, left lower eyelid:right lower eyelid::olfactory sulcus:gingival margin is given as an analogy pair, even though olfactory sulcus and gingival margin are structures in the brain and in the mouth, respectively, and do not share a right:left relationship.[22] Many similar mismatched word pairs were found upon manual inspection, which led to the creation of our novel set of ophthalmology domain-specific analogies, which were all hand-curated and semantically valid. Researchers wishing to evaluate neural word embeddings specific to a particular biomedical domain may also need to develop novel sets of analogies for appropriate testing.
In our ophthalmology analogy set, GloVe general embeddings performed surprisingly well with 91.0% accuracy, outperforming EHR analogies at (86.0%) accuracy. One reason for this may be that all words in the analogy set were limited to vocabulary which was present in all sets of embeddings. We found that while many words in the EHR vocabulary were missing from both GloVe and PubMed embeddings, the most common words missing from GloVe covered important clinical concepts, findings, and tests, whereas the most common words missing from PubMed were less clinically relevant, including more social words like “dear”, “thank”, and “sincerely”. Since analogies had to utilize words common to all vocabulary sets, the highly specialized vocabulary present in the EHR and PubMed which is not covered by GloVe was not tested in the analogies, so the advantages of EHR and PubMed embeddings in greater vocabulary coverage is not reflected in their performance on the analogy test. Thus, differences in vocabulary coverage between different sets of word embeddings must be considered when creating analogy tests, and represents a limitation to their usage as evaluation tools for domain-specific word embeddings. In addition, we found that BioWordVec embeddings actually outperformed all other embeddings on the analogy test, with near-perfect performance. This may be due to the fact that BioWordVec embeddings were trained on an enormous corpus including all PubMed biomedical literature, a superset of the PubMed ophthalmology literature used to train our own ophthalmology-specific domains.
A unique strength of this study was that we also extrinsically evaluated ophthalmology domain-specific word embeddings on a novel clinical task of predicting the visual prognosis of low vision patients using free text clinical notes from the EHR. Many previous studies evaluating medical domain word embeddings use a specific intermediate NLP task, such as named entity recognition, rather than directly on a downstream clinical prediction task, which is an important step for assessing likelihood of success for model deployment.[23–27] Specifically, we were able to predict using free-text EHR notes which visually impaired patients would still be visually impaired one year later, despite ongoing follow-up and treatment, with predictions that substantially outperform a human ophthalmologist with access to the same clinical notes. In the clinical setting, it is important to identify these patients who may most benefit from automated referrals to low vision rehabilitation services. Rather than waiting to observe the effects of treatment before referring to low vision services, an early referral based on predicted prognosis would result in earlier benefits to patients in their quality of life.[6] To our knowledge, there has been no previous work developing machine learning models to predict the visual prognosis for low vision patients, likely due to the lack of a ability to incorporate free-text information, such as afforded by the use of ophthalmology domain-specific word embeddings. Most previous work developing machine learning models in the domain of ophthalmology use imaging data or structured clinical data for classification, diagnosis, and prediction of future outcomes. This includes work predicting progression on visual field testing using imaging data[1,3,28,29], and work predicting glaucoma progression to surgery using structured EHR data.[2] Our method of incorporating ophthalmology free-text notes into deep learning models by using ophthalmology domain-specific word embeddings results in good performance in predicting clinical outcomes. We found that using domain-specific word embeddings resulted in substantial improvements in model performance compared to more general word embeddings, which suggests that training domain-specific word embeddings should be the default approach when planning to use word embeddings to represent highly specialized domain-specific text.
Our approach to developing domain-specific word embeddings and analogy evaluations can serve as an example for those working in other subspecialties with their own highly specialized terminologies, such as obstetrics, neurology, and others, who may wish to boost performance of predictive models by using free text note input features. The pre-trained ophthalmology word embeddings that we have made available can have broad applicability and can be easily used to incorporate EHR free text notes into predictive models for a wide variety of ophthalmology prediction tasks. Furthermore, an advantage of using domain-specific embeddings is that loading and computing over them is likely to require fewer computational resources, owing to their significantly smaller vocabulary (and thus file size) compared to more general vectors. Future refinements to ophthalmology domain-specific word representations can also make use of the novel analogies for benchmarking.
Our approach has several limitations. Word embeddings could only be developed for single words, which does not cover concepts spanning multiple words. Therefore, abbreviations like “amd” would have one embedding vector, whereas the corresponding “age-related macular degeneration” would have separate embeddings for each component word. Analogies were therefore limited also to single words, as well as to words that appeared in all three corpora. The EHRs and clinical notes used in this study were extracted from a single healthcare system, therefore it is possible that we capture local terms and concepts that may not be generalizable across other settings. Other systems may wish to train their own domain-specific embeddings on their own site-specific corpora for the best performance; alternatively, use of our publicly available PubMed-based embeddings could be a ready solution to those not wishing to train their own embeddings as these were trained on ophthalmology literature and would not be expected to exhibit site-specific variation. Additionally, due to our center being a tertiary ophthalmology referral center, a relatively high proportion of our patients have complex or severe eye problems that, while amenable to ongoing treatment, may never recover to a level of vision better than 20/40. Thus, our dataset was fairly balanced, which may not be the case for cohorts of ophthalmology patients seen in other treatment settings. Furthermore, although we were able to predict the visual acuity prognosis of for patients with reduced visual acuity, we recognize that qualifying for low vision services is not merely a question of visual acuity, but also depends on the presence of visual deficits which damage peripheral vision or create specific blind spots while preserving central acuity, as well as functional deficits. Thus, the potential pool of patients who would benefit from low vision rehabilitation services is likely to be larger than the patients we have identified. Future work to improve this model could include identifying these additional patients, combining the free-text unstructured data from clinical progress notes with the structured demographic, diagnosis, medication, and examination data available in the EHR, and experimenting with and the incorporation of the time dimension into predictive models.[30,31]
We also recognize that in recent years, context-aware word embeddings such as those learned in transformer-based approaches[32–34] have grown more popular than embeddings in the style of GloVe or word2vec. These transformer-based models can operate on character-based subwords which can mitigate the issue of out-of-vocabulary words. Future work can experiment with tuning transformer-based models for the ophthalmology domain,[33] and using multiple hierarchical levels of text representation, such as sub-word, paragraph, and/or document-level representations.[35] However, understanding how to customize word2vec type of embeddings is still valuable, as this approach is significantly simpler and computationally less intensive, both to train and to use in model deployment, where resources may be limited. Furthermore, transformer-based approaches with sequence architecture often have additional limitations, such as relatively short limits on the length of text inputs, slower training, and larger data requirements.
In conclusion, we developed novel ophthalmology domain-specific word embeddings using publicly available PubMed ophthalmology literature abstracts as well as EHR ophthalmology notes. We evaluated their performance against more general word embeddings on a novel ophthalmology-specific analogy task as well as on a prediction task using free-text ophthalmology progress notes to predict the visual prognosis of low vision patients. We found that using ophthalmology domain-specific embeddings improved the predictions of deep learning models, suggesting that clinical prediction tasks using highly specialized free text from EHRs benefit from domain-specific word embeddings. Our publicly available ophthalmology word embeddings can be immediately and broadly used for other predictive tasks in ophthalmology using free text clinical progress notes, and our approach can be readily replicated for other subspecialties to improve the performance of other predictive models.
Highlights.
We trained novel word embeddings that are domain-specific to ophthalmology
Ophthalmology word embeddings are now publicly available for further research
We train a deep learning model to predict the visual prognosis of patients with low vision
Performance using ophthalmology embeddings is improved over general word embeddings
SUMMARY POINTS.
What was known:
Neural word embeddings are a powerful way of representing text in general and general biomedical domains.
Publicly available neural word embeddings trained on general English corpora and general biomedical corpora are available for use.
Some medical domains, such as ophthalmology, are highly subspecialized and have unique vocabulary and style not captured in these general corpora.
What this study adds:
Novel word embeddings specifically trained for the highly subspecialized ophthalmology text outperform publicly available off-the-shelf word embeddings in a real-world task of predicting patients’ visual prognosis from information in their electronic health records.
We make ophthalmology domain-specific word embeddings and a set of ophthalmology domain-specific evaluation analogies publicly available for further research use.
Researchers in other subspecialized medical domains with highly unique language may also wish to train subspecialty-specific word embeddings for use in prediction models.
Acknowledgments
FUNDING STATEMENT
Sophia Wang wishes to acknowledge her funding support, NLM T15 LM 007033 to support participation in the Biomedical Informatics training program at Stanford University, as well as a Career Development Award from Research to Prevent Blindness, and unrestricted departmental grants from Research to Prevent Blindness, and the National Eye Institute P30-EY026877.
Footnotes
CONFLICTS OF INTEREST STATEMENT
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Garcia G-GP, Nitta K, Lavieri MS, Andrews C, Liu X, Lobaza E, Van Oyen MP, Sugiyama K, Stein JD, Using Kalman Filtering to Forecast Disease Trajectory for Patients With Normal Tension Glaucoma, Am. J. Ophthalmol. 199 (2019) 111–119. 10.1016/j.ajo.2018.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Baxter SL, Marks C, Kuo T-T, Ohno-Machado L, Weinreb RN, Machine Learning-Based Predictive Modeling of Surgical Intervention in Glaucoma Using Systemic Data From Electronic Health Records, Am. J. Ophthalmol. 208 (2019) 30–40. 10.1016/j.ajo.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wen JC, Lee CS, Keane PA, Xiao S, Rokem AS, Chen PP, Wu Y, Lee AY, Forecasting future Humphrey Visual Fields using deep learning, PLoS One. 14 (2019) e0214875. 10.1371/journal.pone.0214875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ali N, Khan AA, Akunjee M, Ahfat F, Using common ophthalmologic jargon in correspondence can lead to miscommunication, Br. J. Gen. Pract. 56 (2006) 968–969. https://www.ncbi.nlm.nih.gov/pubmed/17132387. [PMC free article] [PubMed] [Google Scholar]
- [5].Hamiel U, Hecht I, Nemet A, Pe’er L, Man V, Hilely A, Achiron A, Frequency, comprehension and attitudes of physicians towards abbreviations in the medical record, Postgrad. Med. J. 94 (2018) 254–258. 10.1136/postgradmedj-2017-135515. [DOI] [PubMed] [Google Scholar]
- [6].Fontenot JL, Bona MD, Kaleem MA, McLaughlin WM Jr, Morse AR, Schwartz TL, Shepherd JD, Jackson ML, American Academy of Ophthalmology Preferred Practice Pattern Vision Rehabilitation Committee, Vision Rehabilitation Preferred Practice Pattern®, Ophthalmology. 125 (2018) P228–P278. 10.1016/j.ophtha.2017.09.030. [DOI] [PubMed] [Google Scholar]
- [7].Coker MA, Huisingh CE, McGwin G Jr, Read RW, Swanson MW, Dreer LE, DeCarlo DK, Gregg L, Owsley C, Rehabilitation Referral for Patients With Irreversible Vision Impairment Seen in a Public Safety-Net Eye Clinic, JAMA Ophthalmol. 136 (2018) 400–408. 10.1001/jamaophthalmol.2018.0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gutiérrez L, Keith B, A Systematic Literature Review on Word Embeddings, in: Trends and Applications in Software Engineering, Springer International Publishing, 2019: pp. 132–141. 10.1007/978-3-030-01171-0_12. [DOI] [Google Scholar]
- [9].Pennington J, Socher R, Manning CD, GloVe: Global Vectors for Word Representation, (2014). https://nlp.stanford.edu/projects/glove/ (accessed June 16, 2020). [Google Scholar]
- [10].Common Crawl, (n.d.). https://commoncrawl.org/ (accessed June 16, 2020).
- [11].Mikolov T, Chen K, Corrado G, Dean J, Efficient Estimation of Word Representations in Vector Space, ArXiv [Cs.CL]. (2013). http://arxiv.org/abs/1301.3781. [Google Scholar]
- [12].Parker R, Graff D, Kong J, Chen K, Maeda K, English gigaword fifth edition, linguistic data consortium, Google Scholar. (2011). [Google Scholar]
- [13].Zhang Y, Chen Q, Yang Z, Lin H, Lu Z, BioWordVec, improving biomedical word embeddings with subword information and MeSH, Sci Data. 6 (2019) 52. 10.1038/s41597-019-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen Q, Peng Y, Lu Z, BioSentVec: creating sentence embeddings for biomedical texts, in: 2019 IEEE International Conference on Healthcare Informatics (ICHI), 2019: pp. 1–5. 10.1109/ICHI.2019.8904728. [DOI] [Google Scholar]
- [15].Lowe HJ, Ferris TA, Hernandez PM, Weber SC, STRIDE--An integrated standards-based translational research informatics platform, AMIA Annu. Symp. Proc. 2009 (2009) 391–395. https://www.ncbi.nlm.nih.gov/pubmed/20351886. [PMC free article] [PubMed] [Google Scholar]
- [16].Wang SY, Pershing S, Tran E, Hernandez-Boussard T, Automated extraction of ophthalmic surgery outcomes from the electronic health record, International Journal of Medical Informatics. 133 (2020) 104007. 10.1016/j.ijmedinf.2019.104007. [DOI] [PubMed] [Google Scholar]
- [17].Wang B, Wang A, Chen F, Wang Y, Kuo C. -C. Jay, Evaluating word embedding models: methods and experimental results, APSIPA Transactions on Signal and Information Processing. 8 (2019). 10.1017/ATSIP.2019.12. [DOI] [Google Scholar]
- [18].Wang Y, Liu S, Afzal N, Rastegar-Mojarad M, Wang L, Shen F, Kingsbury P, Liu H, A comparison of word embeddings for the biomedical natural language processing, J. Biomed. Inform. 87 (2018) 12–20. 10.1016/j.jbi.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang SY, Tseng B, eyelovedata/ophthalmology-wordembeddings: v1.0.1, 2020. 10.5281/zenodo.3932147. [DOI] [Google Scholar]
- [20].Kim Y, Convolutional Neural Networks for Sentence Classification, in: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014: pp. 1746–1751. 10.3115/v1/D14-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang Y, Wallace B, A Sensitivity Analysis of (and Practitioners’ Guide to) Convolutional Neural Networks for Sentence Classification, ArXiv [Cs.CL]. (2015). http://arxiv.org/abs/1510.03820. [Google Scholar]
- [22].Newman-Griffis D, Lai AM, Fosler-Lussier E, Insights into Analogy Completion from the Biomedical Domain, ArXiv [Cs.CL]. (2017). http://arxiv.org/abs/1706.02241. [Google Scholar]
- [23].Sheikhshab G, Birol I, Sarkar A, In-domain Context-aware Token Embeddings Improve Biomedical Named Entity Recognition, in: 2018: pp. 160–164. 10.18653/v1/W18-5618. [DOI] [Google Scholar]
- [24].Liu Y, Ge T, Mathews K, Ji H, McGuinness D, Exploiting Task-Oriented Resources to Learn Word Embeddings for Clinical Abbreviation Expansion, in: 2015: pp. 92–97. 10.18653/v1/W15-3810. [DOI] [Google Scholar]
- [25].Gu Y, Leroy G, Pettygrove S, Galindo MK, Kurzius-Spencer M, Optimizing Corpus Creation for Training Word Embedding in Low Resource Domains: A Case Study in Autism Spectrum Disorder (ASD), AMIA Annu. Symp. Proc. 2018 (2018) 508–517. https://www.ncbi.nlm.nih.gov/pubmed/30815091. [PMC free article] [PubMed] [Google Scholar]
- [26].El Boukkouri H, Ferret O, Lavergne T, Zweigenbaum P, Embedding Strategies for Specialized Domains: Application to Clinical Entity Recognition, in: 2019: pp. 295–301. 10.18653/v1/P19-2041. [DOI] [Google Scholar]
- [27].Jiang Z, Li L, Huang D, Liuke Jin, Training word embeddings for deep learning in biomedical text mining tasks, in: 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), ieeexplore.ieee.org, 2015: pp. 625–628. 10.1109/BIBM.2015.7359756. [DOI] [Google Scholar]
- [28].Berchuck SI, Mukherjee S, Medeiros FA, Estimating Rates of Progression and Predicting Future Visual Fields in Glaucoma Using a Deep Variational Autoencoder, Sci. Rep. 9 (2019) 18113. 10.1038/s41598-019-54653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang M, Shen LQ, Pasquale LR, Petrakos P, Formica S, Boland MV, Wellik SR, De Moraes CG, Myers JS, Saeedi O, Wang H, Baniasadi N, Li D, Tichelaar J, Bex PJ, Elze T, An Artificial Intelligence Approach to Detect Visual Field Progression in Glaucoma Based on Spatial Pattern Analysis, Invest. Ophthalmol. Vis. Sci. 60 (2019) 365–375. 10.1167/iovs.18-25568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Banerjee I, Gensheimer MF, Wood DJ, Henry S, Aggarwal S, Chang DT, Rubin DL, Probabilistic Prognostic Estimates of Survival in Metastatic Cancer Patients (PPES-Met) Utilizing Free-Text Clinical Narratives, Sci. Rep. 8 (2018) 10037. 10.1038/s41598-018-27946-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Banerjee I, Bozkurt S, Caswell-Jin JL, Kurian AW, Rubin DL, Natural Language Processing Approaches to Detect the Timeline of Metastatic Recurrence of Breast Cancer, JCO Clin Cancer Inform. 3 (2019) 1–12. 10.1200/CCI.19.00034. [DOI] [PubMed] [Google Scholar]
- [32].Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, Levy O, Lewis M, Zettlemoyer L, Stoyanov V, RoBERTa: A Robustly Optimized BERT Pretraining Approach, ArXiv [Cs.CL]. (2019). http://arxiv.org/abs/1907.11692. [Google Scholar]
- [33].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J, BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics. 36 (2020) 1234–1240. 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sanh V, Debut L, Chaumond J, Wolf T, DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter, ArXiv [Cs.CL]. (2019). http://arxiv.org/abs/1910.01108. [Google Scholar]
- [35].Yang Z, Yang D, Dyer C, He X, Smola A, Hovy E, Hierarchical attention networks for document classification, in: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, aclweb.org, 2016: pp. 1480–1489. https://www.aclweb.org/anthology/N16-1174.pdf. [Google Scholar]