ABSTRACT
Nowadays, extracellular DNA or circulating cell-free DNA is considered to be a molecule with clinical applications (diagnosis, prognosis, monitoring of treatment responses, or patient follow-up) in diverse pathologies, especially in cancer. Nevertheless, because of its molecular characteristics, it can have many other functions. This review focuses on the participation of extracellular DNA (exDNA) in fundamental processes such as cell signaling, coagulation, immunity, evolution through horizontal transfer of genetic information, and adaptive response to inflammatory processes. A deeper understanding of its role in each of these processes will allow development of better tools to monitor and control pathologies, as well as helping to generate new therapeutic options, beyond the applicability of DNA in liquid biopsy.
KEYWORDS: Extracellular DNA, second messenger DNA, horizontal DNA transfer, oxidized DNA, extracellular DNA traps
Introduction
It is a common knowledge that in eukaryotes, DNA is located inside the cell nucleus. One of the first studies that reported the presence of DNA in the extracellular space, specifically in plasma, was performed by the French researchers Mandel and Métais in 1948;1 they demonstrated the presence of DNA in the plasma of patients and healthy individuals. Extracellular DNA (exDNA) has been observed in many conditions such as inflammatory and autoimmune diseases, lupus erythematosus, myocardial infarction, diabetes, and cancer.2 Subsequent studies suggested that this exDNA acts as a reflection of the disease state so it has been a subject of study ever since. In this finding, some concepts were defined: exDNA which refers to DNA located beyond cellular membrane; while circulating DNA (circDNA) refers to the exDNA molecules found in circulating physiological fluids such as bloodstream, lymphatic system, bile, milk, urine, saliva, mucous suspension, spinal fluid, and amniotic fluid.3,4
Regarding the biology of exDNA, the most studied aspects have been the origin of exDNA and the release mechanisms. exDNA originates from all cells and it can be present in the circulation in different forms, mainly in macromolecular complexes (linked with proteins, lipids, or other nucleic acids) or associated with extracellular vesicles.5 This largely depends on the way the exDNA is released from the cell, and a variety of passive and active exDNA release mechanisms have been described in the literature. These studies have demonstrated that the release of DNA to the extracellular space might occur 1) directly by metabolically active secretion of living cells through macromolecular structures such as DNA-protein complexes (1000 to 3000 base pairs) and DNA extracellular traps,4 by micro-enucleation of extrachromosomal circular DNA induced by genomic instability (30 to 20,000 bp of size) and through vesicular transport (exosomes, virtosome, and AGO2),6–8 or 2) indirectly as a consequence of the mechanisms of cell disruption generated by the different death pathways including apoptosis and necrosis;9–13 as well as other pathways comprising neutrophil extracellular trap release, phagocytosis, and oncosis.14–16 Processes that increase the release of exDNA include disease, inflammation, tissue injury, and exercise.5 In healthy individuals, hematopoietic maturation is a major contributor to the normal exDNA pool. Multiple studies since then further confirmed that the lymphoid/myeloid tissues mainly contribute to the normal exDNA pool.17
In cancer patients, the level of exDNA was estimated to be threefold higher compared with healthy individuals.18 Diehl et al. reported that the exDNA level correlates with tumor burden, that is to say, the amount of exDNA in a patients’ blood is closely correlated with tumor burden and increases significantly with tumor growth and/or with disease progression.19 However, it has been postulated that circulating tumor cells are not the only source of total exDNA; thus, it can be speculated that another part of the exDNA also originates from cells in the tumor microenvironment, from cells involved in the antitumor response, and potentially from stressed normal cells.20
Concerning the clinical applications of exDNA in plasma/serum or biological fluid, most studies have been done in the oncological field. Early studies demonstrated that the DNA released to the extracellular space retains the genetic and epigenetic characteristics of the tissue from which it was released, for example, changes in DNA integrity,21 mutations in oncogenes or tumor suppressor genes, gene methylation abnormalities, microsatellite alterations,22 changes in mitochondrial DNA load levels,23 chromosomal genome rearrangements, etc.22 Additionally, the concentration in blood can reflect the latest developments and specificity of tumors in real time.24 Due to these characteristics, exDNA was proposed as a possible cancer biomarker.25 Currently, the diagnostic importance of exDNA is increasingly noticeable; in fact, its importance in oncology has led to thousands of studies within the novel field of “liquid biopsy”. In short, liquid biopsy is a noninvasive diagnostic test that provides information on early cancer detection, accurate diagnosis, therapy response, prognosis, and follow-up avoiding the invasive procedures needed to obtain tissue samples, particularly when these are needed serial samples.26 The treatment of some cancer types has improved because of increased knowledge of the molecular abnormalities that drive human cancer growth. This has led to the development of targeted cancer therapies.27 In light of these advances, the testing of molecular biomarkers for cancer patient stratification has become mandatory. Such tests are performed routinely using biopsy/cytology material from primary tumors obtained at the time of diagnosis. While this approach is suitable for diagnostic purposes, it precludes the follow-up of the patient during disease progression and eventual relapse. The concept of liquid biopsy was introduced to oncology with the potential to revolutionize the management of cancer patients, eliminating the invasive procedures needed to obtain tissue samples and provide information on therapy response and disease relapse on the fly.28–30 Currently, liquid biopsy is already used in clinical trials, there are, however, some hurdles that need to be overcome in order to introduce liquid biopsy into routine clinical practice.31–34 This interesting issue is beyond the scope of this review.
However, the biological role of exDNA in the organisms remains uncertain. In this review, we are trying to explore some biological processes as ç cellular signaling, oxidative stress response, in inflammation or innate immunity, blood coagulation, and evolution through horizontal transfer of genetic information in which exDNA could participate directly or indirectly.
Role of exDNA in cellular signaling
In 1969, E. Bell identified for the first time an association between exDNA and intercellular communication, through DNA molecules packaged in lipoprotein complexes named informational DNA (I-DNA) or informasome (I-somes). This I-DNA is nuclear DNA (not mitochondrial) found in the cytoplasm, described as an information intermediary between the nucleus and cytoplasm acting as a template for protein synthesis.35–37
It has been suggested that exDNA is a biologically active molecule and could be categorized as a heterogeneous complex entity made up of different types of DNA: free DNA fragments, vesicle bound DNA, and DNA-macromolecular complexes (proteolipid nucleic acid complexes), which are formed by the electrostatic and autocondensation properties of DNA and whose function is to provide protection against nucleases and immune surveillance.4,20,38–40 The exDNA can have different activities, either when it enters the cell or when it interacts with the surface of the target cells, acting as an intercellular messenger thus triggering various biological responses; the nucleotide sequence is not always implicit in its functions.4,40–42 Recent studies have proposed that the specific signaling properties of exDNA partly depend on diverse factors including sequence length, 3D structure, the association of DNA molecules with histones and non-histone chromatin-binding, subcellular localization, methylation status, sequence (i.e. dinucleotide CG content), and oxidation status (8-oxodG).20,43–46
The exDNA free or associated with macromolecules (proteins-lipids), is detected by sensors in the cell membrane of leucocytes4,47 and by ligand-receptors in the cell cytoplasm, e.g., RIG-1, AIM2, DAI, IFI16, and STING43 which are part of the family PRRs (Patterns Recognition Receptors) and that can recognize to exDNA as DAMPs (Damage-Associated Molecular Patterns).48 It has also been reported that some proteins such as albumin are able to form complexes with exDNA and favor its internalization into the cell via endocytosis.20 Once inside the cell, exDNA is detected by TLR-9 (Toll-like receptor 9) located in the endosome membrane and is activated by unmethylated sequences.49–55
On the other hand, the DNA bound to extracellular vesicles (EVs) is biologically functional and serves like an intercellular messenger, and can regulate the biological functions of the target cells by increasing DNA-coding mRNA and protein levels stimulating their proliferation and induce phenotype changes.16,38 The EVs contain DNA fragments randomly selected from the entire genome spanning the 5ʹ promoter region, gene coding region, and 3ʹ untranslated region of chromatin-associated double-stranded DNA.38 The exDNA is transported either on the surface or inside diverse EVs predominantly apoptotic bodies, microvesicles, and exosomes,56 the latter being the most related to cell–cell communication that results in the activation of signaling pathways such as the activation of the interferon type I (IFN-I) response through the cGAS-STING;20 and eliciting molecular responses in the target cells.56 The exosomes can be transferred cell-to-cell through different mechanisms such as endocytosis, fusion, or through specific receptor binding.38 It appears that exosomes may use several types of receptors to facilitate its internalization either directly, e.g., C-type lectin, CD33, cadherin 11, integrin α6β4, CD9, CD81, and TIM1/TIM4, or indirectly, e.g., EGFR.57
The horizontal transfer of genomic DNA and the signal transduction might mediate intercellular communication and influence the functions of the affected cells, either by integrating into the genome or binding to receptors of target cells to elicit a biological effect, such as induction of tolerance against detrimental substances, immunological changes, development of metastasis, and generation of genetic instability that enables transformation to cancer cells.20,36,39,40,47 This type of activity of exDNA (free or bound-EVs) has been observed in cardiomyopathies, cancer, Alzheimer’s disease, and skeletal muscle diseases, acting by intercellular communication, thus influencing the biological functions of recipient cells.38
A possible activity of exDNA in cell signaling was tested in vitro: T cells were extracted from donors and then exposed to Herpes Simplex Virus (HSV); T cells’ released DNA following stimulation by HSV was isolated and added to B cells culture for 3 days resulting in the synthesis of anti-HSV antibodies. The same was observed in in vivo assays when nude mice were injected with the DNA released in the culture of T cells exposed to HSV or polioviruses; 5 days after injection, the nude mice produced specific anti-HSV or anti-polio antibodies without having been directly exposed to the mentioned pathogens. These experiments seem to indicate that the T cell released DNA could act as a mediator in cell signaling involved in the adaptive immune response.4,47
Role of exDNA in oxidative stress response
To keep homeostasis either at the molecular, cellular, or tissular level, different pathways are activated. Kostyuk and collaborators have demonstrated that oxidized exDNA influences survival and cell death, depending on the type of receptor cell that acquires the DNA; their model suggests that oxidized exDNA stimulates different pathways implicated in the inflammatory response depending on the translocation of NF-kB to the nucleus, which favors the synthesis of pro-inflammatory cytokines such as TNFα, IL-1, IL-2, IL-6, IL-12, and adhesion molecules.58 This increases the levels of Reactive Oxygen Species (ROS) being the effect in the immune response stimulation 12 times greater in comparison with non-oxidized exDNA.58 The magnitude of the response differs greatly since non-oxidized exDNA causes a weak and prolonged response of oxidative stress due to the increase in the expression of NF-kB and NRF-2, which favors chronic long-term cell death.58,59 8-oxodG triggers a massive and acute cell death by inducing a reduction in the production of nitric oxide (NO) along with the reduction of endothelial NO-synthase60, increasing NAD(P)H oxidases (coded by NOX4).61 Oxidative stress decreases proliferation in fibroblasts associated with a decrease in Ki-67, subsequently causing cell death.58,60
On the other hand, the activity of systems that protect cells from ROS, for instance, NRF-2, is increased by the effect of the oxidized exDNA in mesenchymal stem cells, meanwhile, in cell lines like HUVEC and MCF-7, the levels of NF-kB and NOX4 are increased instead.48,61 All this indicates that GC content in DNA as well as its oxidation level are directly involved in cell survival.58,59
It is widely accepted that ROS produced during oxidative stress cause damage to nuclear and mitochondrial DNA; therefore, ROS released to the extracellular medium could oxidize exDNA to form 8-oxodG,triggering a feedback cycle between the production of 8-oxodG and its internalization in cells, causing a greater production of ROS and so on until the cell dies.58,62 Based on this, it has been suggested that oxidized exDNA might be an oxidative stress marker and that its level of oxidation is associated with the abundance of the CG dinucleotide due to its susceptibility to be oxidized.63 The presence of this exDNA has been linked to the inhibition of proliferation and to ROS-triggered apoptosis.63,64
In this regard, a recent study demonstrated that oxidized exDNA (8-oxodG) is a signaling molecule that regulates the radio-adaptive response through a “bystander” effect in cells adjacent to the irradiation site.62 The bystander effect has been linked to development of genomic instability, cell death, and adaptive responses by the bystander cells, which in turn depend on the irradiation-dose received. Radiation itself triggers primary oxidative stress, increasing the levels of ROS, causing damage to genomic DNA by the rupture of the deoxyribose rings and consequently, the appearance of apuric/apyrimidinic sites, double-strand breaks, DNA-protein cross-linking, and the formation of oxidized bases.62,65 All this increases 8-oxodG levels, induce apoptosis and favors the release of oxidized exDNA, which is then acquired by the bystander cells and triggers the pathways involved in the initial steps of apoptosis. However, during the adaptive response, the increase in intracellular ROS comes accompanied by the activation of antioxidant responses that lead to the repair of DNA rupture which in turn explains the cell cycle' arrest in the short term.62,66,67 On the other hand, low doses of radiation might induce a radio-adaptive response and the inhibition of apoptosis due to the reduction of ROS levels.65
In conclusion, the binding of (dG)n oxidized exDNA to a variety of receptors triggers different signaling pathways, as has been observed with the binding to TLR-9 which activates NF-kB and increases the levels of ROS. To a certain extent, whether DNA is repaired, or cell death is undergone, depends on the differentiation state of each cell (Figure 1).58,65
Figure 1.
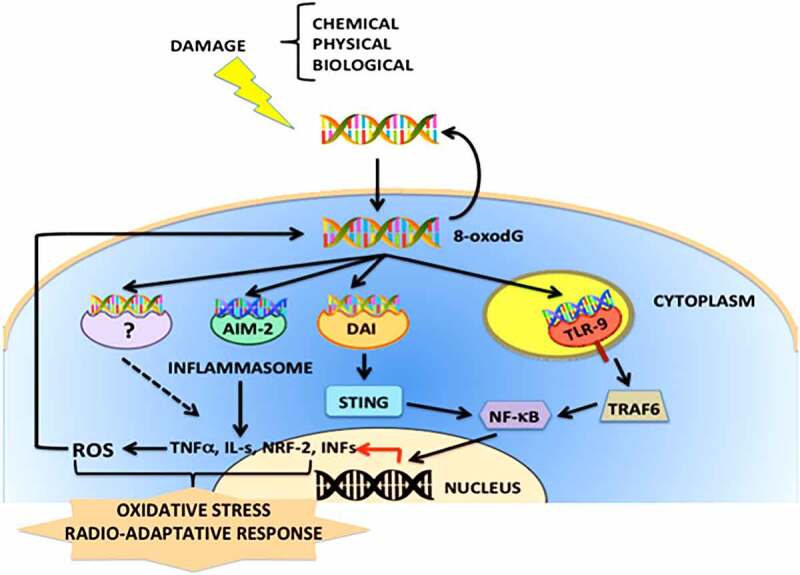
Schematic representation of the cellular signaling pathways activated by oxidized exDNA (8-oxodG). The exDNA released by different mechanisms of cellular stress and cell damage, might be enriched with oxidized nucleotides, forming complexes with proteins and/or lipids. The 8-oxodG acting as DAMP (Damage-Associated Molecular Pattern) signal might easily enter into cells and interact with different PRRs (Patterns Recognition Receptors) such as STING, AIM-2, RIG-1 or DAI, and ROS (Reactive Oxygen Species) induces DDR (DNA Damage Response) responses
Role of exDNA in inflammation (or innate immunity)
Different studies have shown the relationship between exDNA and the immune system, since exDNA can be recognized by TLR-9,68,69 which triggers the production of cytokines, specifically those involved in inflammatory processes, and activates neutrophils which might release DNA extracellular traps, known as Neutrophil Extracellular Traps (NETs).70,71 However, this phenomenon is not exclusive to neutrophils since it has been reported that extracellular DNA traps might also be released by eosinophils (EETs), basophils (BETs), mast cells, and plant cells.72,73 It has also been observed that activated T cells, B cells, NK cells, and monocytes are also able to release mitochondrial DNA forming extracellular web-like structures.73
NETs are extracellular structures that resemble networks consisting of DNA, histones, granules, cytoplasmic proteins, neutrophil elastase, myeloperoxidase, cathepsin G, proteinase 3, gelatinase, LL-37, lactoferrin, and calprotectin. NETs’ main function is to trap microorganisms to avoid dissemination.74,75 This supramolecular structure contains a high local concentration of granulocytic enzymes released by neutrophils, that eliminates the invading microorganism (bacteria, fungi, viruses, and parasites).73 The mechanism that initiates the formation of NETs is known as NETosis and might be triggered by the Pathogen-Associated Molecular Patterns (PAMPs) as well as by inflammatory cytokines (TNF and IL-8), and by the chromatin binding protein HMGB1.76 This mechanism is also combined with oxidative stress, since ROS is required to initiate NETosis, which induces actin and tubulin glutathionylation. Therefore, an intact cytoskeleton is required for the formation of NETs.73,75 During this process genomic DNA is uncoiled by the peptidyl arginine-deiminase 4 (PAD4), a nuclear enzyme that converts arginine residues into citrulline in histones 3 and 4; with this, the positive charge of the histones is neutralized favoring chromatin decondensation.77,78 NETs also activate the coagulation cascade, which contributes to the antimicrobial activity of NETs. The combined action of these factors results in the antimicrobial activity of NET´s. In summary, the antimicrobial activity of NETs comes from the combined action of all the components that make it up.72,79
Histones are essential for eukaryotes and archaea; however, in the middle of the last century some researchers found that histones are potent antibiotics80 that can eliminate bacteria at concentrations in the nanomolar order.81 In mammals, the antimicrobial activity or NET is exerted through the formation of ROS resulting from the enzymatic activity of different proteins, by the slight alkalinization of the area (pH 7.6), and by direct contact with the antimicrobial peptides found immersed within a network made up of naked DNA and histones (polynucleosomes).75 The histones released to the extracellular space through NETosis have been found in the cell surface of the affected organ, interacting with receptors such as TLR-2, TLR-4 y TLR-9,75,82 having cytotoxic effects in epithelial and endothelial cells in a dose-dependent way.83 The binding of histones to the cellular membrane might be due to its cationic nature, this could cause an increase in the cell permeability through the formation of holes in the membrane.84 It has also been demonstrated that extracellular histones can bind to the plasmid DNA of prokaryotes84 interfering with the activity of DNA-gyrase.85 Moreover, the cytotoxic activity of these extracellular histones in NETs during the inflammation process can also kill the eukaryotic cells, classifying them as an essential component in the toxicity observed in the sepsis process.83,86,87
On the other hand, the innate immune system has evolved to detect and react to the interruption of the systemic homeostasis; disturbances are sensed through the PAMP and DAMP receptors.88 In the case of exDNA, it is detected by extra and intracellular receptors, such as TLR-9, which resides inside the phagosomes of monocytes and dendritic cells. TLR-9 is preferentially activated by GpC enriched-unmethylated DNA, which is more abundant in prokaryotes than eukaryotes.79 It has also been demonstrated that exDNA bound to the antimicrobial LL-37 (cathelicidin antimicrobial peptide of 37 amino acids) or to the HMGB1 protein activates dendritic cells more potently than naked DNA, and this is because exDNA bound to different molecules forms stable structures, which ensure recognition and interaction with various receptors like TLR-9.79,89
Recently, it has been pointed out that exDNA can bind to IL-26 due to its cationic charge, facilitating its entrance to the receptor cell using two alpha-helices with amphipathic characteristics contained in its sequence as an anchor to the membrane.90 Binding of exDNA to IL-26 involves the activation of STING, a transmembrane protein that might be directly activated by DNA or by intermediary cytosolic sensors like cGAS (cyclic GMP-AMP synthase). The complex exDNA-IL 26 induces the production of IL 6 and IL 1β in human monocytes.90 Based on this evidence, the authors conclude that IL-26 confers immuno-stimulating properties to different types of DNA (genomic, mitochondrial, and NETs) which triggers the secretion of proinflammatory cytokines in monocytes and NK cells.54
In addition, it has been shown that NETs also have a critical role in noninfectious pathologies, such as 1) systemic lupus erythematous, characterized by the synthesis of autoantibodies against self-DNA and antimicrobial peptides present in NETs that trigger B-cell activation, 2) small vessel vasculitis, where NETs cause the expansion of Th1 pathogenic cells through maturation of dendritic cells leading to the production of IFN-γ, 3) atherosclerosis, in which it has been demonstrated that NETs can stimulate plasmacytoid dendritic cells to produce IFN-α, promoting atherosclerotic plaque growth, and 4) cancer, being tumor cells the ones that cause the stimulation of neutrophils, through the release of IL-8 and G-CSF to form NETs, creating a favorable microenvironment for metastasis. In summary, the presence of NETs in this type of disease finally leads to the vicious cycle of chronic inflammation sterile.73
This kind of immunity mediated by NETs is no exclusive to mammals, it has also been reported that the roots of several plants actively release DNA; this was demonstrated when the roots were fed with the radioactive phosphorus labeled dCTP which was collected in the extracellular medium without evident cell death.91,92 This extracellular nucleic acid form structures similar to NETs; although until now the main DNA release mechanism is not known, it is evident that its function must be similar to mammals since these plant-networks also avoid the invasion of pathogenic microorganisms on the root of the plant (Figure 2).91,92
Figure 2.
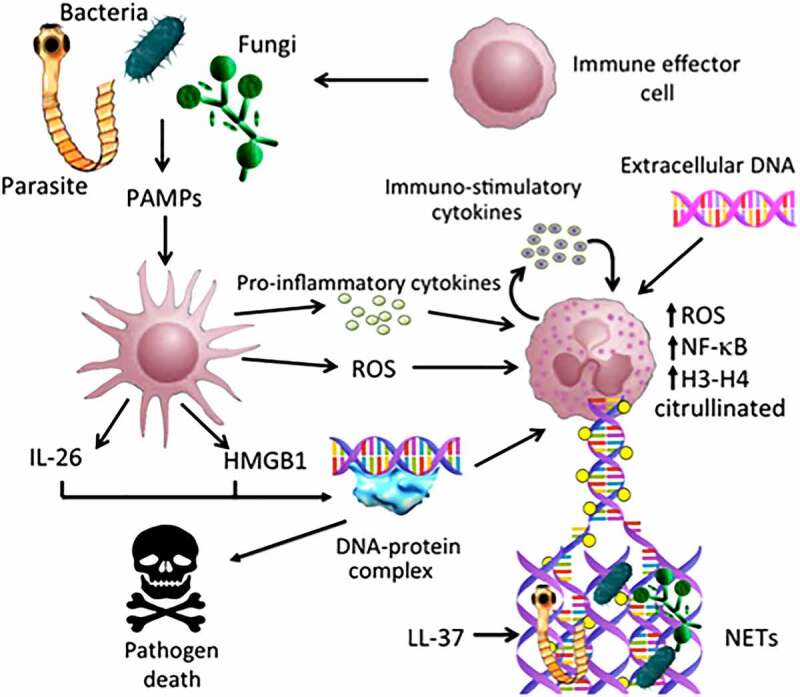
Possible implications of exDNA in immune response. Bacteria, fungi and other opportunistic parasites might be recognized by the immune system cells such as macrophages and dendritic cells, and thus induce the release of pro-inflammatory cytokines as well as the generation of ROS (Reactive Oxygen Species). Several stimuli trigger the activation and set up of the NETs (Neutrophil Extracellular Traps), which involve the participation of several proteins such as PAD4, NF-kB and histones, that help in the construction of the DNA molecular network and to trap different pathogens. All this induces the participation of multiple immune cells, which ultimately leads to the death and elimination of exogenous microorganisms
In the case of cancer, Albrengues showed that proliferative disseminated breast cancer cells during inflammation process induced by smoking, the activated NETs, initiate the awakening of dormant metastatic cells.93 The DNA along with MMP-9, integrin α3β 1, FAK (focal adhesion kinase) and laminin-111 plays an important role in the evasion of immune system and cancer progression.93,94
Role of exDNA in blood coagulation
Recently, the participation of proteins that belong to the coagulation cascade in the release of chromatin has been reported, particularly Factor VII Activating Protease (FSAP) which has been identified as responsible for nucleosome release.95 FSAP associates with the DNAse I participating in the degradation of DNA, in the nucleosome release in apoptotic and necrotic cells, and physically binding them by an unknown mechanism; forming complexes with its targets serpin α2-antiplasmin (AP) and the C1 inhibitor (C1-inh). FSAP might be activated by nucleosomes, glycosaminoglycans, and RNA, probably through the release of its inhibitors.96,97
A recent analysis has revealed that exDNA is considered as a “natural singular surface” of the human body that promotes blood clotting in vivo. Evidence that support this asseveration, is that purified genomic DNA increases the activation of proteases that participate in the contact pathway of blood clotting, such as the coagulation factors XII (FXII) and XI (FXI); similarly, it has been observed that the exDNA from activated neutrophils that are part of the NETs, trigger blood clotting that relies on FXII and FXI.98,99 This was corroborated through a scanning with an electron microscope, which demonstrated that exDNA binds to FXII and HK (high molecular weight kininogen) and that the double-stranded structure with highly negative charge density of the exDNA are required to promote procoagulant activity given by a high affinity for HK.100,101 Furthermore, it has been observed that histones interact with the A1 domain of the von Willebrand human factor (VWF), which can propagate platelet adhesion mediated by GPIbα.102
NETs have been shown to interbreed with the fibrin clot forming a structural network resistant to DNA lysis and to the tissue plasminogen-activator (tPA), due to the addition of the chromatin-bound to fibrin, the formation of thicker and steadier fibers which are more resistant to shear forces is induced.103 These findings indicate that histones modify the structure of the clot holding the fibrillar network together. An in vitro study showed that fibrin co-localized with the exDNA in the clot.103 Consisting with the above, a recent study revealed that histones can bind to platelets and recruit plasma adhesion proteins, such as fibrinogen, causing platelet aggregation inducing thrombotic occlusion of the microvasculature and thrombocytopenia in an in vivo model; this finding suggests that the extracellular interaction of histones with glycosaminoglycans negatively charged and located on the surface of endothelial cells, might contribute to the activation of clotting and interfere with the anticoagulant properties of the endothelium´s glycocalyx.104 Likewise, it has been determined that the increase in the concentrations of exDNA increases the viscosity of blood, which causes problems with microcirculation since autoimmune immunogenic processes are triggered.72
Role of in horizontal genetic transfer
It has been clearly demonstrated that exDNA has the ability to enter other cells in vitro and in vivo, being capable of transferring genetic information. Consequently, the biology of the recipient cells is modified since the exDNA contains sequences and even mutations similar to their parent cells, and this might result in inherited modifications.105 The first report that suggested this ability of the exDNA was done by Perc106 in 1968 who named this transferable DNA as “metabolic DNA”. This fact was later corroborated by Rogers107 in 1972 and by Anker108 3 years later. In parallel, in the year 1965, it was hypothesized that the exDNA might be involved in the spread of metastases, due to its ability to travel in the bloodstream and/or the lymphatic system.109 Years later, García-Olmo et al.,110 called this phenomenon “genometastasis” and in 2010 demonstrated that the exDNA found in the plasma of cancer patients transfers oncogenic information to susceptible cells in vitro,110 concluding that metastasis can occur through the transfection of susceptible cells in distant organs with the sequences of dominant oncogenes that circulate in plasma. Due to these characteristics, exDNA obtained from cancer patients was called “oncosome”, which has also been related to the promotion of angiogenesis.111,112
The non-clonal transference of exDNA has been corroborated through the malignant transformation of the cell line NIH3T3 (immortalized non-neoplastic murine fibroblasts) after the incubation of the cells with the serum of colon cancer patients, strongly suggesting that the DNA contained in the serum might be acquired and integrated by the receptor cells.113,114 This event can also occur in nonpathogenic conditions such as pregnancy; the transference and integration of a specific segment of the Y chromosome in the brain of a woman who gave birth to a male fetus has been observed. This description gave rise to the phenomenon called “microchimerism” derived from the horizontal gene transfer.115
According to the previous information, Mittra and collaborators in 2015 showed for the first time that the exDNA from patients and healthy individuals, which was transferred to NHI3T3 for later specific identification of fragments of the exDNA in the nucleus of NIH3T3, might induce apoptosis and DNA ruptures in vitro and in vivo.116 It should be noted that the DNA extracted from the sera of patients and healthy individuals was fragments of chromatin and DNA of apoptotic cells. However, this work only focused on the damage generated but never considered the possible functional effects, such as changes in the cellular metabolism or changes in the overall enzyme activity.116
An important feature of exDNA that might permit its entrance to the target cells, is the fact that it contains a great number of (dG)n motifs. Particularly, human cells have GpC enriched sequences in nuclear DNA, with a large number of (dG)n motifs, specifically in repetitive ribosomal sequences; in fact, this characteristic allows to differentiate between mitochondrial and bacterial DNA. The length of these sequences might be (dG)11 or (dG)13 and when oxidized, they are able to increase adaptive responses up to 20 times.48 It has also been observed that exDNA sequences that contain (dG)n motifs can enter the cell nucleus and be expressed, allowing cell survival in the case of cancer cells.48,59 It has been postulated that the genomic DNA of cancer cells has a rich content of these amplified sequences as well as of activated oncogenes.117,118 The amplified sequences might have copies of certain chromosomal regions. The exDNA can also have amplified sequences, favoring the synthesis of certain proteins when it enters cells.119,120 The presence of transposable mobile genetic elements in exDNA can facilitate the transference of fragments of DNA from one cell to another, as well as the illicit integration into the genome of the receptor cells, contributing to genome instability.119,121
A recent study demonstrated that when exDNA enters a cell, it is recognized by the receptor TLR-9; initiating the NF-kB signaling pathway which leads to an increment in the expression of cyclin D through the TLR signaling axis TLR9/NK-kB/Cyclin D1. This promoted proliferation of at least one hormone-dependent breast cancer cell line.40 Other authors have confirmed that exDNA is released by cells that can act as an intercellular messenger, having two possibilities when it enters a receptor cell: 1) to integrate into the host genome or 2) to bind to certain receptors and trigger different signaling pathways that ultimately lead to some biological effects, e.g., induction of tolerance toward detrimental substances, immunomodulation, metastasis development or genomic instability.40,119,122
In agreement with the above, Tuomela and collaborators in 2013123 demonstrated that the exDNA from doxorubicin-killed MDA-MB-231 cells could promote invasion of this same wild type cell line when exposed to the cell debris and that this event was mediated by the receptor TLR-9.123 Likewise, García-Arranz124 concluded that the exDNA from non-neoplastic cells can reduce cell proliferation and metastasis; which suggests that the released exDNA might have different effects depending on the biological context of the cell from which it is originated. Using these effects as a possible method of inhibiting tumor growth.124,125
Perspectives
In past years, liquid biopsy has involved an active research field, in the attention to early cancer detection.126 The use of circulating tumor DNA in accurate diagnosis, prognosis determination, treatment selection, and serial monitoring of disease is enormously crucial in the context of survival, quality life, and response to treatment in cancer patients.127 The use of this information can be employed for a personalized depiction of the disease; having other advantages: is noninvasive, its low cost, its adequate and efficient.126,127
However, the biological role of exDNA has another scope not only in cancer research, but also in several medical applications, taken remarkable steps toward other pathologies and normal physiological conditions. The data presented in this review propose that exDNA has functions beyond what has been historically established (see Table 1); suggesting that this biomolecule associated with proteins and lipids when is actively secreted, serves as a molecular messenger between neighboring cells; possibly to synchronize the activities of the cells or as a collective reservoir of lost functions, which could be easily recovered by the incorporation of this DNA when physiological conditions demand it. In agreement with the above, the release of exDNA among the different biological kingdoms suggests that chromatin evolved with two different functions: 1) to organize large DNA fragments and 2) to act as a defense mechanism maintaining the genome integrity. A possible example of chromatin as a safeguard and regulator of genetic information is the structures known as NETs, which could be one of the configurations where chromatin is used to guard against foreign organisms. However, exDNA is not only a component of chromatin or NETs but it can also activate the immune system through different receptors like TRL-9, TLR-2, and TLR-4 which induce the recruitment of different cells to trigger an adaptive immune response or to dissolve an inflammatory process.
Table 1.
Biological processes in which exDNA participates in eukaryotes
| Biological process | exDNA type | Key molecules | Chemical or biological characteristics | Probable biological effect |
|---|---|---|---|---|
| Cell signaling | Associated with lipoprotein complexes | Receptors: STING, AIM-2 RIG-1, DAI, TLRs | High content of 8-oxodG | Second messengersExchange of informationbetween nucleus and cytoplasmintra- and intercellular |
| Oxidative stress | Oxidized | NF-κB, TNF α, IL-1, 2, 6 & 12NRF-2, adhesion molecules | High content of 8-oxodG | Keeping of homeostasis |
| Radio-adaptiveresponses | Oxidized andnon-oxidized | TLR-9, NF-κB | Receptor cell biology | Cell cycle arrestAnti-oxidative response |
| Immunity | Chromatin | TLR-9, PAMPs, HMGB1, ROS, PAD4, IL-26, LL-37, Pro inflammatory cytokines, | Citrullination of histones H3 and H4 | NETs formation Antimicrobial activity |
| Coagulation | Nucleosome | FSAP, Factor XI– XII, HK, VWF, GPIbα | Presence of NETs, glycosaminoglycan and RNA | Activation of the cascade Thrombus formation Increased blood viscosity |
| Gene transference | Naked DNA Chromatin Oncosome Apoptotic bodies | TRL-9, NF-κB, cyclin D1 | Susceptible receptor cell | Inheritable biological modifications Angiogenesis Metastasis and invasion Microchimerism Apoptosis |
| Gene amplification | small fragments | Transposase? | Presence of transposable gene elements | Gain of metabolic functions Genomic instability |
On the other hand, the acquisition of exogenous DNA has a role in evolution, since it provides favorable characteristics in the development and survival of organisms in their habitats, facilitating the acquisition of genes and genetic polymorphisms among prokaryotes or eukaryotes and between them. The importance of horizontal gene transference is hard to determine, but it is clear that it has a transcendental role in the microbial evolution; it has been estimated that between 1.6% and 32% of the genes in the genomes of these organisms have been acquired in this way.128 In the case of eukaryotes, the acquisition of new genes is also a driver in their evolutionary process as seen clearly in the case of endosymbiosis and in genetic hybridization.128
In recent years, there has been increasing evidence that complexes of DNA associated with proteins and lipids are more effective than naked DNA in gene delivery to the nucleus, which raises the following questions: 1) How exDNA travels during the pathophysiological processes of different diseases? And what is the difference with that released under normal conditions? 2) Is this exDNA a linear structure with or without DNA binding proteins? The knowledge of the composition of other components that interact with exDNA will provide a better understanding of the homeostasis of extracellular nucleic acids and the different interaction with several target cells.
New approaches to using exDNA as a therapeutic target in the treatment of different human diseases have been proposed. For example, Linardou in 1995 suggested the employed of recombinant DNAse I in cancer therapy.129 DNAse I and DNAse1L3 are endonucleases involved in repair, replication, and degradation of DNA, and in the homeostasis of exDNA.130,131 The treatment with DNAse I in in vitro and in vivo cancer models reduces the tumor cell proliferation, migration, adhesion, and invasion and correlates a significant reduction of levels of plasma exDNA and metastasis development.93,130,132 In patients with cancer it has been reported a decrease in DNAse blood levels with a concomitant exDNA increment.130 Part of the therapeutic effect of DNAse is mediated by the inhibition of NETosis and the anti-inflammatory and immunomodulatory reactions.133 Alzheimer´s disease is another possibility for the use of DNAse I.134 The systemic administration of this endonuclease can affect the amyloid cascade and the rupture or fusion of exosomes, suggesting three possible mechanisms of action: an anti-inflammatory effect, the dissolution of NETs, and the reduction of DNA-amyloid-β complex.134,135
Finally, the efforts made so far have shown a plausible overview of the biological role of exDNA in cellular homeostasis. However, it is still evident the need for more adequate tools and biological models that help to understand the functionality of exDNA, and the pathological changes associated directly with the presence of exDNA.
Acknowledgments
This work was supported by CONACyT under Grant No. 6901; CONACyT grant no. 465479. This work is submitted in partial fulfillment of the requirements for the PhD degree (in Biological Sciences) of Ileana Jocelyn Fernández-Domínguez at the Universidad Nacional Autónoma de México.
Funding Statement
This work was supported by the Consejo Nacional de Ciencia y Tecnología grants [6901].
Disclosure of interest
The authors report no conflict of interest.
References
- 1.Mandel P, Métais P.. Les acides nucléiques du plasma sanguin chez l’homme [The nucleic acids of blood plasma in humans]. C R Seances Soc Biol Fil. 1948;142(3–4):241–243. [PubMed] [Google Scholar]
- 2.Aarthy R, Mani S, Velusami S, Sundarsingh S, Rajkumar T. 2015. Role of circulating cell-free DNA in cancers. Mol Diagn Ther. 19(6):339–350. 10.1007/s40291-015-0167-y. [DOI] [PubMed] [Google Scholar]
- 3.Gahan PB, Anker P, Stroun SM. 2008. Metabolic DNA as the origin of spontaneously released DNA?. Ann NY Acad Sci. 1137(1):7–17. 10.1196/annals.1448.046. [DOI] [PubMed] [Google Scholar]
- 4.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. 2016. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 35(3):347–376. 10.1007/s10555-016-9629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zukowski A, Rao S, Ramachandran S. 2020. Phenotypes from cell-free DNA. Phenotypes from cell-free DNA. Open Biol. 10(9):200119. 10.1098/rsob.200119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rykova E, Morozkin E, Ponomaryova A, Loseva E, Zaporozhchenko I, Cherdyntseva N, Vlassov V, Laktionov P. 2012. Cell-free and cell-bound circulating nucleic acid complexes: mechanisms of generation, concentration and content. Expert Opin Biol Ther. 12(Suppl sup1):S141–S153. 10.1517/14712598.2012.673577. [DOI] [PubMed] [Google Scholar]
- 7.Marzban C, Viswanathan R, Yurtsever U. 2014. Earth before life. Biol Direct. 9(11):1–11. 10.1186/1745-6150-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. 2017. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes?. Cell Physiol Biochem. 41(2):755–768. 10.1159/000458736. [DOI] [PubMed] [Google Scholar]
- 9.Lichtenstein A, Melkonyan H, Tomei LD, Umansky SR. Circulating nucleic acids and apoptosis. Ann NY Acad Sci. 2006;945(1):239–249. doi: 10.1111/j.1749-6632.2001.tb03892.x. tb03892.x.. [DOI] [PubMed] [Google Scholar]
- 10.Stroun M, Lyautey J, Olson-Sand A, Anker P. 2001. About the possible origin and mechanism of circulating DNA. Apoptosis and active DNA release. Clin Chim Acta. 313(1–2):139–142. 10.1016/s0009-8981(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 11.Ulivi P, Silvestrini R. 2013. Role of quantitative and qualitative characteristics of free circulating DNA in the management of patients with non-small cell lung cancer. Cell Oncol. 36(6):439–448. 10.1007/s13402-013-0155-3. [DOI] [PubMed] [Google Scholar]
- 12.Bryzgunova OE, Laktionov PP. 2014. Generation of blood circulating DNAs: sources, features of structure and circulation. Biochemistry (Moscow) Supplement Series B: Biomed Chem. 8(3):203–219. 10.1134/S1990750814030020. [DOI] [Google Scholar]
- 13.Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. 2018. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc. 93(3):1649, 168. 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 14.Bronkhorst AJ, Wentzel JF, Aucamp J, Van Dyk E, Du Plessis L, Pretorius PJ. 2016. Characterization of the cell-free DNA released by cultured cancer cells. Biochim Biophys Acta. 1863(1):157–165. 10.1016/j.bbamcr.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;614:1659–1665. PMID: 11245480. [PubMed] [Google Scholar]
- 16.Pös O, Biró O, Szemes T, Nagy B. 2018. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 26(7):937–945. 10.1038/s41431-018-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam WKJ, Gai W, Sun K, Wong RSM, Chan RWY, Jiang P, Chan NPH, Hui WWI, Chan AWH, ChCh S, et al.. 2017. DNA of erythroid origin is present in human plasma and informs the types of anemia. Clin Chem. 63:1614–1623. 10.1373/clinchem.2017.272401. [DOI] [PubMed] [Google Scholar]
- 18.Sato Y, Matoba R, Kato K. 2019. Recent advances in liquid biopsy in precision oncology research. Biol Pharm Bull. 42(3):337–342. 10.1248/bpb.b18-00804. [DOI] [PubMed] [Google Scholar]
- 19.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al.. 2008. Circulating mutant DNA to assess tumor dynamics. Nat Med. 14(9):985–990. DOI: 10.1038/nm.1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther. 2019;20(8):1057–1067. doi: 10.1080/15384047.2019. 1598759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umetani N, Giuliano AE, Hiramatsu SH, Amersi F, Nakagawa T, Martino S, Hoon DS. 2006. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol. 24(26):4270–4276. 10.1200/JCO.2006.05.9493. [DOI] [PubMed] [Google Scholar]
- 22.Ludovini V, Pistola L, Gregorc V, Floriani I, Rulli E, Piattoni S, Di Carlo L, Semeraro A, Darwish S, Tofanetti FR, et al.. 2008. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: a study of the Perugia multidisciplinary team for thoracic oncology. J Thorac Oncol. 3(4):365–373. DOI: 10.1097/JTO.0b013e318168c7d0 [DOI] [PubMed] [Google Scholar]
- 23.Li L, Hann HW, Wan S, Hann RS, Wang C, Lai Y, Ye X, Evans A, Myers RE, Ye Z, et al.. 2016. Cell-free circulating mitochondrial DNA content and risk of hepatocellular carcinoma in patients with chronic HBV infection. Sci Rep. 6(1):23992. DOI: 10.1038/srep23992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierry AR, Mouliere F, Gongora C, Ollier J, Robert B, Ychou M, Del Rio M, Molina F. 2010. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res. 38(18):6159–6175. 10.1093/nar/gkq421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duforestel M, Briand J, Bougras-Cartron G, Heymann D, Frenel J-S, Vallette FM, Cartron P-F. 2020. Cell-free circulating epimarks in cancer monitoring. Epigenomics. 12(2):145–155. 10.2217/epi-2019-0170. [DOI] [PubMed] [Google Scholar]
- 26.Fernández-Lázaro D, García Hernández JL, García AC, Córdova Martínez A, Mielgo-Ayuso J, Cruz-Hernández JJ. 2020. Liquid Biopsy as Novel Tool in Precision Medicine: origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics (Basel). 10(4):215. 10.3390/diagnostics10040215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. 2020. Molecular profiling for precision cancer therapies. Genome Med. 12(1):8. 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz LA, Bardelli BA. 2014. Liquid Biopsies: genotyping Circulating Tumor DNA. J Clin Oncol. 32(6):579–586. 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen SØ, Øgaard N, Ørntoft MBW, Rasmussen MH, Bramsen JB, Kristensen H, Mouritzen P, Madsen MR, Madsen AH, Sunesen KG, et al.. 2019. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer—a clinical biomarker discovery and validation study. Clin Epigenetics. 11(1):158. DOI: 10.1186/s13148-019-0757-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warton K, Mahon KL, Samimi G. 2016. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer. 23(3):R157–R171. 10.1530/ERC-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, Gray SW, Goetz L, Goel A, Schork N, et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol Ther. 2020; 207(107458). doi: 10.1016/j.pharmthera.2019.107458. pharmthera.2019.107458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Q, Zhang Z-H, Wang S, Lang J-H. 2019. <p>Circulating Cell-Free DNA or Circulating Tumor DNA in the Management of Ovarian and Endometrial Cancer. Onco Targets Ther. 12:11517–11530. 10.2147/OTT.S227156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Chen P, Xie H, Cao P. 2020. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: a systematic review and meta-analysis. Cancer Med. 9(4):1349–1364. 10.1002/cam4.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reece M, Saluja H, Hollington P, Karapetis CS, Vatandoust S, Young GP, Symonds EL. 2019. The use of circulating tumor DNA to monitor and predict response to treatment in colorectal cancer. Front Genet. 10(1118):1–25. 10.3389/fgene.2019.01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell E. 1969. I-DNA: its packaging into I-somes and its relation to protein synthesis during differentiation. Nature. 224(5217):326–328. 10.1038/224326a0. [DOI] [PubMed] [Google Scholar]
- 36.Aucamp J, Bronkhorst AJ, Badenhorst CP, Pretorius PJ. 2016. A historical and evolutionary perspective on the biological significance of circulating DNA and extracellular vesicles. Cell Mol Life Sci. 73(23):4355–4381. 10.1007/s00018-016-2370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell BE. 1971. Informational DNA Synthesis Distinguished from That of Nuclear DNA by Inhibitors of DNA Synthesis. Science. 174(4009):603–606. 10.1126/science.174.4009.603. [DOI] [PubMed] [Google Scholar]
- 38.Cai J, Wu G, Jose PA, Zeng C. 2016. Functional transferred DNA within extracellular vesicles. Exp Cell Res. 349(1):179–183. 10.1016/j.yexcr.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Gahan PB, Stroun M. 2010. The virtosome-a novel cytosolic informative entity and intercellular messenger. Cell Biochem Funct. 28(7):529–538. 10.1002/cbf.1690. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Kong P, Ma G, Li L, Zhu J, Xia T, Xie H, Zhou W, Wang S. 2017. Characterization of the release and biological significance of cell-free DNA from breast cancer cell lines. Oncotarget. 8(26):43180–43191. 10.18632/oncotarget.17858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagler M, Insam H, Pietramellara G, Ascher-Jenull A-J-J. 2018. Extracellular DNA in natural environments: features, relevance and applications. Appl Microbiol Biotechnol. 102(15):6343–6356. 10.1007/s00253-018-9120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otandault A, Anker P, Al Amir DZ, Guillaumon V, Meddeb R, Pastor B, Pisareva E, Sanchez C, Tanos R, Tousch G, et al.. 2019. Recent advances in circulating nucleic acids in oncology. Ann Oncol. 30(3):374–384. DOI: 10.1093/annonc/mdz031 [DOI] [PubMed] [Google Scholar]
- 43.Vanpouille-Box C, Demaria S, Formenti SC, Galluzzi GL. 2018. Cytosolic DNA Sensing in Organismal Tumor Control. Cancer Cell. 34(3):361–378. 10.1016/j.ccell.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Lehmann-Werman R, Neiman D, Zemmour H, Moss J, Magenheim J, Vaknin-Dembinsky A, Rubertsson S, Nellgård B, Blennow K, Zetterberg H, et al.. 2016. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci USA. 113(13):E1826–E1834. DOI: 10.1073/pnas.1519286113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skvortsova TE, Rykova EY, Tamkovich SN, Bryzgunova OE, Starikov AV, Kuznetsova NP, Vlassov VV, Laktionov PP. 2006. Cell-free and cell-bound circulating DNA in breast tumours: DNA quantification and analysis of tumour-related gene methylation. Br J Cancer. 94(10):1492–1495. 10.1038/sj.bjc.6603117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veĭko NN, Bulycheva NV, Roginko OA, Veĭko RV, Ershova ES, Kozdoba OA, Kuz’min VA, Vinogradov AM, Iudin AA, Speranskiĭ AI. 2008. [Ribosomal repeat in the cell free DNA as a marker for cell death]. Biomed Khim. 54(1):78–93. 10.1134/S1990750808020121. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z, Fadiel A, Naftolin F, Eichenbaum KD, Xia XY. 2005. Circulation DNA: biological implications for cancer metastasis and immunology. Med Hypotheses. 65(5):956–961. 10.1016/j.mehy.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 48.Kozhina EA, Ershova ES, Okorokova NA, Veiko VP, Malinovskaya EM, Sergeeva VA, Konkova MS, Kutsev SI, Veiko NN, Kostyuk SV. 2019. Extracellular DNA Containing (dG)n Motifs Penetrates into MCF7 Breast Cancer Cells, Induces the Adaptive Response, and Can Be Expressed. Oxid Med Cell Longev. 2019:7853492. 10.1155/2019/7853492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unterholzner L. 2013. The interferon response to intracellular DNA: why so many receptors?. Immunobiology. 218(11):1312–1321. 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34(5):637–650. doi: 10.1016/j.immuni.2011.05.006. 05.006. [DOI] [PubMed] [Google Scholar]
- 51.Harberts E, Gaspari AA. 2013. TLR signaling and DNA repair: are they associated?. J Invest Dermatol. 133(2):296–302. 10.1038/jid.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 243(1):91–98. 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 53.Keating SE, Baran M, Bowie AG. 2011. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 32(12):574–581. 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 54.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 339(6121):786–791. 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 339(6121):826–830. 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kalluri R, LeBleu VS. 2016. Discovery of double-stranded genomic DNA in circulating exosomes. Cold Spring Harbor Symposia on Quantitative Biology. 81:275–280. 10.1101/sqb.2016.81.030932. [DOI] [PubMed] [Google Scholar]
- 57.Gonda A, Kabagwira J, Senthil GN, Wall NR. 2019. Internalization of exosomes through receptor-mediated endocytosis. Mol Cancer Res. 17(2):337–347. 10.1158/1541-7786.MCR-18-0891. [DOI] [PubMed] [Google Scholar]
- 58.Kostyuk SV, Porokhovnik LN, Ershova ES, Malinovskaya EM, Konkova MS, Kameneva LV, Dolgikh OA, Veiko VP, Pisarev VM, Martynov AV, et al.. Changes of KEAP1/NRF2 and IKB/NF-κB expression levels induced by cell-free DNA in different cell types. Oxid Med Cell Longev. 2018;1052413. doi: 10.1155/2018/1052413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kostyuk SV, Konkova MS, Ershova ES, Alekseeva AJ, Smirnova TD, Stukalov SV, Kozhina EA, Shilova NV, Zolotukhina TV, Markova ZG, et al.. 2013. An exposure to the oxidized DNA enhances both instability of genome and survival in cancer cells. PLoS One. 8(10):e77469. DOI: 10.1371/journal.pone.0077469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kostyuk SV, Alekseeva AY, Kon’kova MS, Glebova KV, Smirnova TD, Kameneva LV, Izhevskaya VL, Veiko NN. 2014. Oxidized extracellular DNA suppresses nitric oxide production by endothelial NO synthase (eNOS) in human endothelial cells (HUVEC). Bull Exp Biol Med. 157(2):202–206. 10.1007/s10517-014-2525-x. [DOI] [PubMed] [Google Scholar]
- 61.Loseva P, Kostyuk S, Malinovskaya E, Clement N, Dechesne CA, Dani C, Smirnova T, Glebova K, Baidakova G, Baranova A, et al.. 2012. Extracellular DNA oxidation stimulates activation of NRF2 and reduces the production of ROS in human mesenchymal stem cells. Expert Opin Biol Ther. 12(1):S85–S97. DOI: 10.1517/14712598.2012.688948 [DOI] [PubMed] [Google Scholar]
- 62.Konkova MS, Kaliyanov AA, Sergeeva VA, Abramova MS, Kostyuk SV. 2019. Oxidized cell-free DNA is a factor of stress signaling in radiation-induced bystander effects in different types of human cells . International Journal of Genomics. 2019:9467029. 10.1155/2019/9467029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valavanidis A, Vlachogianni T, Fiotakis C. 2009. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. Journal of Environmental Science and Health, Part C. 27(2):120–139. 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 64.Roszkowski K, Jozwicki W, Blaszczyk P, Mucha-Malecka A, Siomek A. 2011. Oxidative damage DNA: 8-oxoGua and 8-oxodG as molecular markers of cancer. Med Sci Monit. 17(6):CR329–CR333. 10.12659/msm.881805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sergeeva VA, Ershova ES, Veiko NN, Malinovskaya EM, Kalyanov AA, Kameneva LV, Stukalov SV, Dolgikh OA, Konkova MS, Ermakov AV, et al.. 2017. Low-dose ionizing radiation affects mesenchymal stem cells via extracellular oxidized cell-free DNA: a possible mediator of bystander effect and adaptive response. Oxid Med Cell Longev. 2017:9515809. 10.1155/2017/9515809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ermakov AV, Konkova MS, Kostyuk SV, Izevskaya VL, Baranova A, Veiko NN. 2013. Oxidized extracellular DNA as a stress signal in human cells. Oxid Med Cell Longev. 2013:649747. 10.1155/2013/649747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glebova K, Veiko N, Kostyuk S, Izhevskaya V, Baranova A. 2015. Oxidized extracellular DNA as a stress signal that may modify response to anticancer therapy. Cancer Lett. 356(1):22–33. 10.1016/j.canlet.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 68.Guiducci C, Coffman RL, Barrat FJ. 2009. Signalling pathways leading to IFN-α production in human plasmacytoid dendritic cell and the possible use of agonists or antagonists of TLR7 and TLR9 in clinical indications. J Intern Med. 265(1):43–57. 10.1111/j.1365-2796.2008.02050.x. [DOI] [PubMed] [Google Scholar]
- 69.Nathan C. 2006. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 6(3):173–182. 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 70.Papayannopoulos V, Zychlinsky A. 2009. NETs: a new strategy for using old weapons. Trends Immunol. 30(11):513–521. doi: 10.1016/j.it.2009.07.011.. [DOI] [PubMed] [Google Scholar]
- 71.Kaplan MJ, Radic M. 2012. Neutrophil extracellular traps: double-edged swords of innate immunity. J Immunol. 189(6):2689–2695. 10.4049/jimmunol.1201719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liaw PC, Ito T, Iba T, Thachil J, Zeerleder ZS. 2016. DAMP and DIC: the role of extracellular DNA and DNA-binding proteins in the pathogenesis of DIC. Blood Rev. 30(4):257–261. 10.1016/j.blre.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Yousefi S, Simon D, Stojkov D, Karsonova A, Karaulov A, Simon H-U. In vivo evidence for extracellular DNA trap formation. Cell Death Dis. 2020;11(4):300. Published 2020 April30. doi: 10.1038/s41419-020-2497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daniel C, Leppkes M, Muñoz LE, Schley G, Schett G, Extracellular HM. 2019. DNA traps in inflammation, injury and healing. Nat Rev Nephrol. 15(9):559–575. 10.1038/s41581-019-0163-2. [DOI] [PubMed] [Google Scholar]
- 75.Lou H, Pickering MC. 2018. Extracellular DNA and autoimmune diseases. Cell Mol Immunol. 15(8):746–755. 10.1038/cmi.2017.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tadie J-M, Bae H-B, Jiang S, Park DW, Bell CP, Yang H, Pittet J-F, Tracey K, Thannickal VJ, Abraham E, et al.. 2013. HMGB1 promotes neutrophil extracellular trap formation through interactions with Toll-like receptor 4. Am J Physiol Lung Cell Mol Physiol. 304(5):L342–9. DOI: 10.1152/ajplung.00151.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neeli I, Dwivedi N, Khan S, Radic M. 2009. Regulation of extracellular chromatin release from neutrophils. J Innate Immun. 1(3):194–201. 10.1159/000206974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Li M, Stadler S, Correll S, Li P, Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, et al.. 2009. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 184(2):205–213. DOI: 10.1083/jcb.200806072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brinkmann V, Zychlinsky A. 2012. Neutrophil extracellular traps: is immunity the second function of chromatin?. J Cell Biol. 198(5):773–783. 10.1083/jcb.201203170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller BF, Abrams R, Dorfman A, Klein M. 1942. Antibacterial properties of protamine and histone. Science. 96(2497):428–430. 10.1126/science.96.2497.428. [DOI] [PubMed] [Google Scholar]
- 81.Hirsch JG. 1958. Bactericidal action of histone. J Exp Med. 108(6):925–944. 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, Brinkmann V, Jungblut PR, Zychlinsky A, Levitz SM. 2009. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5(10):e1000639. 10.1371/journal.ppat.1000639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saffarzadeh M, Juenemann C, Queisser MA, Lochnit G, Barreto G, Galuska SP, Lohmeyer J, Preissner KT, Hartl D. 2012. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PLoS One. 7(2):e32366. 10.1371/journal.pone.0032366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawasaki H, Koyama T, Conlon JM, Yamakura F, Iwamuro S. 2008. Antimicrobial action of histone H2B in Escherichia coli: evidence for membrane translocation and DNA-binding of a histone H2B fragment after proteolytic cleavage by outer membrane proteinase T. Biochimie. 90(11–12):1693–1702. 10.1016/j.biochi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 85.Lemaire S, Trinh -T-T, Le H-T, Tang S-C, Hincke M, Wellman-Labadie O, Ziai S. 2008. Antimicrobial effects of H4-(86-100), histogranin and related compounds - possible involvement of DNA gyrase. Febs J. 275(21):5286–5297. 10.1111/j.1742-4658.2008.06659.x. [DOI] [PubMed] [Google Scholar]
- 86.Gupta AK, Joshi MB, Philippova M, Erne P, Hasler P, Hahn S, Resink TJ. 2010. Activated endothelial cells induce neutrophil extracellular traps and are susceptible to NETosis-mediated cell death. FEBS Lett. 584(14):3193–3197. 10.1016/j.febslet.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 87.Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, Taylor FB, Esmon NL, Lupu F, Esmon CT. 2009. Extracellular histones are major mediators of death in sepsis. Nat Med. 15(11):1318–1321. 10.1038/nm.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pisetsky DS. 2012. The origin and properties of extracellular DNA: from PAMP to DAMP. Clin Immunol. 144(1):32–40. 10.1016/j.clim.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang Y-H, Homey B, Cao W, Wang Y-H, Su B, Nestle FO, et al.. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 449(7162):564–569. DOI: 10.1038/nature06116 [DOI] [PubMed] [Google Scholar]
- 90.Poli C, Augusto JF, Dauvé J, Adam C, Preisser L, Larochette V, Pignon P, Savina A, Blanchard S, Subra JF, et al.. 2017. IL-26 confers proinflammatory properties to extracellular DNA. J Immunol. 198(9):3650–3661. DOI: 10.4049/jimmunol.1600594 [DOI] [PubMed] [Google Scholar]
- 91.Hawes MC, Curlango-Rivera G, Wen F, White GJ, Vanetten HD, Xiong XZ. 2011. Extracellular DNA: the tip of root defenses?. Plant Sci. 180(6):741–745. 10.1016/j.plantsci.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 92.Wen F, White GJ, VanEtten HD, Xiong Z, Hawes MC. 2009. Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol. 151(2):820–829. 10.1104/pp.109.142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, Upadhyay P, Uyeminami DL, Pommier A, Küttner V, et al.. 2018. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science. 361(eaao4227):1–13. DOI: 10.1126/science.aao4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aguirre-Ghiso JA. 2018. How dormant cancer persists and reawakens. Science. 361(6409):1315. 10.1126/science.aav0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zeerleder S, Zwart B, Te Velthuis H, Stephan F, Manoe R, Rensink I, Aarden LA. 2008. Nucleosome-releasing factor: a new role for factor VII-activating protease (FSAP). Faseb J. 22(12):4077–4084. 10.1096/fj.08-110429. [DOI] [PubMed] [Google Scholar]
- 96.Stephan F, Hazelzet JA, Bulder I, Boermeester MA, Van Till JW, Van Der Poll T, Wuillemin WA, Aarden LA, Zeerleder S. 2011. Activation of factor VII-activating protease in human inflammation: a sensor for cell death. Crit Care. 15(2):R110. 10.1186/cc10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stephan F, Marsman G, Bakker LM, Bulder I, Stavenuiter F, Aarden LA, Zeerleder S. 2014. Cooperation of factor VII-activating protease and serum DNase I in the release of nucleosomes from necrotic cells. Arthritis Rheumatol. 66(3):686–693. 10.1002/art.38265. [DOI] [PubMed] [Google Scholar]
- 98.Swystun LL, Mukherjee S, Liaw PC. 2011. Breast cancer chemotherapy induces the release of cell-free DNA, a novel procoagulant stimulus. J Thromb Haemost. 9(11):2313–2321. 10.1111/j.1538-7836.2011.04465.x. [DOI] [PubMed] [Google Scholar]
- 99.Massberg S, Grahl L, Von Bruehl M-L, Manukyan D, Pfeiler S, Goosmann C, Brinkmann V, Lorenz M, Bidzhekov K, Khandagale AB, et al.. 2010. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat Med. 16(8):887–896. DOI: 10.1038/nm.2184 [DOI] [PubMed] [Google Scholar]
- 100.Oehmcke S, Mörgelin M, Herwald H. 2009. Activation of the human contact system on neutrophil extracellular traps. J Innate Immun. 1(3):225–230. 10.1159/000203700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gansler J, Jaax M, Leiting S, Appel B, Greinacher A, Fischer S, Preissner KT. 2012. Structural requirements for the procoagulant activity of nucleic acids. PLoS ONE. 7(11):e50399(1–10). 10.1371/journal.pone.0050399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ward CM, Tetaz TJ, Andrews RK, Berndt MC. 1997. Binding of the von Willebrand factor A1 domain to histone. Thromb Res. 86(6):469–477. 10.1016/s0049-3848(97)00096-0. [DOI] [PubMed] [Google Scholar]
- 103.Fuchs TA, Brill A, Wagner DD. 2012. Neutrophil extracellular trap (NET) impact on deep vein thrombosis. Arterioscler Thromb Vasc Biol. 32(8):1777–1783. 10.1161/ATVBAHA.111.242859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chaaban H, Keshari RS, Silasi-Mansat R, Popescu NI, Mehta-D’Souza P, Lim YP, Lupu F. 2015. Inter-α inhibitor protein and its associated glycosaminoglycans protect against histone-induced injury. Blood. 125(14):2286–2296. 10.1182/blood-2014-06-582759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Keeling PJ, Palmer JD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 9(8):605–618. 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 106.Pelc SR. 1968. Turnover of DNA and function. Nature. 219(5150):162–163. 10.1038/219162a0. [DOI] [PubMed] [Google Scholar]
- 107.Rogers JC, Boldt D, Kornfeld S, Skinner A, Valeri CR. 1972. Excretion of deoxyribonucleic acid by lymphocytes stimulated with phytohemagglutinin or antigen. Proc Natl Acad Sci U S A. 69(7):1685–1689. 10.1073/pnas.69.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Anker P, Stroun M, Maurice PA. Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 1975;35(9):2375–2382. Published September 1975. [PubMed] [Google Scholar]
- 109.Bendich A, Wilczok T, Circulating BE. 1965. DNA as a possible factor in oncogenesis. Science. 148(3668):374–376. 10.1126/science.148.3668.374. [DOI] [PubMed] [Google Scholar]
- 110.García-Olmo DC, Domínguez C, García-Arranz M, Anker P, Stroun M, García-Verdugo JM, García-Olmo D. 2010. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 70(2):560–567. 10.1158/0008-5472.CAN-09-3513. [DOI] [PubMed] [Google Scholar]
- 111.Jaiswal R, Sedger LM. Intercellular vesicular transfer by exosomes, microparticles and oncosomes – implications for cancer biology and treatments. Front Oncol. 2019;9:125. Published 2019 March6. doi: 10.3389/fonc.2019.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Minciacchi VR, Freeman MR, Di Vizio D. 2015. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 40:41–51. 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trejo-Becerril C, Pérez-Cárdenas E, Dueñas-González A. 2014. In vivo rat model to study horizontal tumor progression. Methods Mol Biol. 1165:175–185. 10.1007/978-1-4939-0856-1_12. [DOI] [PubMed] [Google Scholar]
- 114.Trejo-Becerril C, Pérez-Cárdenas E, Taja-Chayeb L, Anker P, Herrera-Goepfert R, Medina-Velázquez LA, Hidalgo-Miranda A, Pérez-Montiel D, Chávez-Blanco A, Cruz-Velázquez J, et al.. 2012. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS One. 7(12):e52754. DOI: 10.1371/journal.pone.0052754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan WF, Gurnot C, Montine TJ, Sonnen JA, Guthrie KA, Nelson JL. 2012. Male microchimerism in the human female brain. PLoS One. 7(9):e45592. 10.1371/journal.pone.0045592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mittra I, Khare NK, Raghuram GV, Chaubal R, Khambatti F, Gupta D, Gaikwad A, Prasannan P, Singh A, Iyer A, et al.. 2015. Circulating nucleic acids damage DNA of healthy cells by integrating into their genomes. J Biosci. 40(1):91–111. DOI: 10.1007/s12038-015-9508-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beljanski M, Bourgarel P, Beljanski M. 1981. Correlation between in vitro DNA synthesis, DNA strand separation and in vivo multiplication of cancer cells. Exp Cell Biol. 49(4):220–231. 10.1159/000163825. [DOI] [PubMed] [Google Scholar]
- 118.Xing R, Zhou Y, Yu J, Yu Y, Nie Y, Luo W, Yang C, Xiong T, Wu WKK, Li Z, et al.. 2019. Whole-genome sequencing reveals novel tandem-duplication hotspots and a prognostic mutational signature in gastric cancer. Nat Commun. 10(1):2037. DOI: 10.1038/s41467-019-09644-6x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gravina S, Sedivy JM, Vijg J. 2016. The dark side of circulating nucleic acids. Aging Cell. 15(3):398–399. 10.1111/acel.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mukherjee K, Storici F. 2012. A mechanism of gene amplification driven by small DNA fragments. PLoS Genet. 8(12):e1003119. 10.1371/journal.pgen.1003119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schaack S, Gilbert C, Promiscuous FC. 2010. DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 25(9):537–546. 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Volik S, Alcaide M, Morin RD, Cell-free CC. 2016. DNA (cfDNA): clinical significance and utility in cancer shaped by emerging technologies. Mol Cancer Res. 14(10):898–908. 10.1158/1541-7786.MCR-16-0044. [DOI] [PubMed] [Google Scholar]
- 123.Tuomela J, Sandholm J, Kaakinen M, Patel A, Kauppila JH, Ilvesaro J, Chen D, Harris KW, Graves D, Selander KS. 2013. DNA from dead cancer cells induces TLR9-mediated invasion and inflammation in living cancer cells. Breast Cancer Res Treat. 142(3):477–487. 10.1007/s10549-013-2762-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garcia-Arranz M, Garcia-Olmo D, Vega-Clemente L, Stroun M, Preliminary A. 2017. Study of the action of virtosomes from non-dividing cells on tumour cell replication in vitro and in vivo. Anticancer Agents Med Chem. 17(10):1401–1410. 10.2174/1871520617666170213110536. [DOI] [PubMed] [Google Scholar]
- 125.García-Olmo D, Garcia-Arranz M, Clemente LV, GahanPB SM. 2015. Method for blocking tumour growth. European Patent 2015. Application number 13169783.1 (EP 2 808 027 A1).
- 126.Fiala C, Diamandis E. 2018. Utility of circulating tumor DNA in cancer diagnostics with emphasis on early detection . BMC Med. 16(166):1–10. 10.1186/s12916-018-1157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heitzer E, Haque IS, Roberts CE, Speicher MR. 2019. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nature Rev Genetics. 20:71–88. 10.1038/s4576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 128.Boto L. 2010. Horizontal gene transfer in evolution: facts and challenges. Proc Biol Sci. 277(1683):819–827. 10.1098/rspb.2009.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Linardou H, Decnarain MP, Epenetos AA. 1995. Recombinant deoxyribonuclease I (DNAse I) and chimeras in cancer therapy. Eur J Cancer. 31(6):s30. . [DOI] [Google Scholar]
- 130.Hawes MC, Wen F, Extracellular EE. 2015. DNA: a bridge to cancer. Cancer Res. 75(20):4260–4264. 10.1158/0008-5472.CAN-15-1546. [DOI] [PubMed] [Google Scholar]
- 131.Serpas L, Chan RWY, Jiang P, Ni M, Sun K, Rashidfarrokhi A, Soni C, Sisirak V, Lee WS, Cheng SH, et al.. 2019. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc Natl Acad Sci U S A. 116(2):641–649. DOI: 10.1073/pnas.1815031116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alcázar-Leyva S, Cerón E, Masso F, Montaño LF, Gorocica P, Alvarado-Vásquez N. Incubation with DNase I inhibits tumor cell proliferation. Med Sci Monit. 2009;15(2):CR51–55. [PubMed] [Google Scholar]
- 133.Mutua V, Gershwin LJ. A review of neutrophil extracellular traps (NETs) in disease: potential anti-NETs therapeutics. Clin Rev Allergy Immunol. 2020;1–18. doi: 10.1007/s12016-020-08804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Smalheiser NE. 2019. Mining clinical case reports to identify new lines of investigation in Alzheimer´s disease: the curious case of DNAse I. J Alzheimers Dis Rep. 3(1):71–76. 10.3233/ADR-190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stefanius K, Servage K, De Souza Santos M, Gray HF, Toombs JE, Chimalapati S, Kim MS, Malladi VS, Brekken R, Orth K. 2019. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. Elife. 8:e40226. 10.7554/eLife.40226. [DOI] [PMC free article] [PubMed] [Google Scholar]


