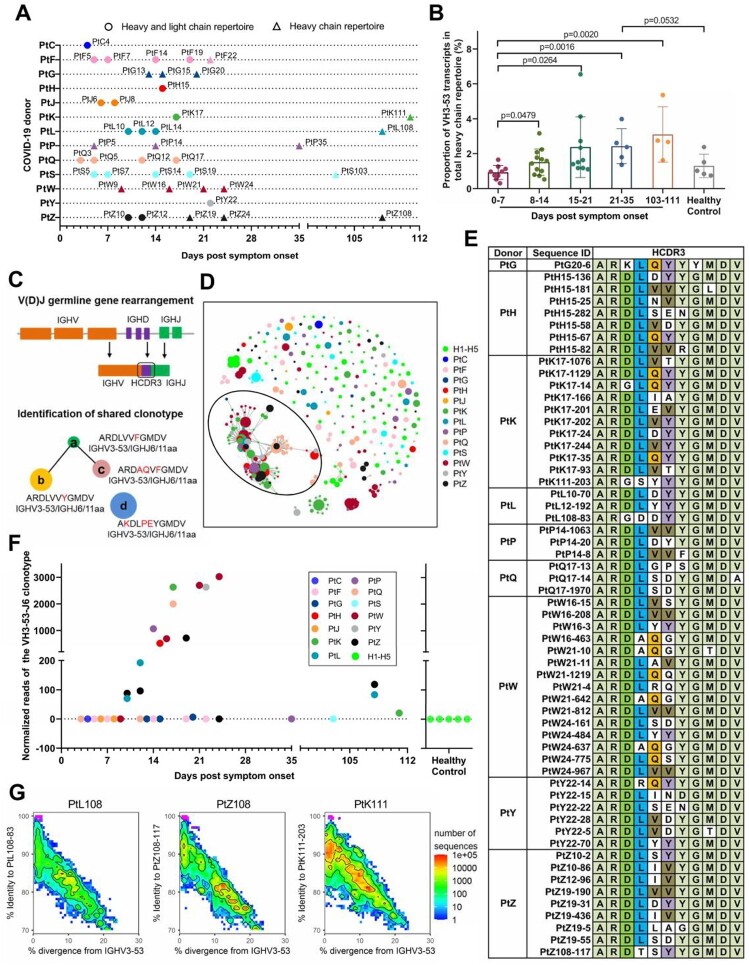
Figure 1.
Shared VH3-53-derived antibodies were observed in individuals exposed to SARS-CoV-2. (A) Scheme of longitudinal sample collection and antibody repertoire sequencing. Circles indicate samples used for both heavy- and light-chain repertoire sequencing, while triangles indicate samples used for only heavy-chain repertoire sequencing. (B) Comparison of VH3-53 germline gene usage in heavy chain repertoires between different time points after the onset of symptoms. Statistical tests were performed by Student’s t-test (Unpaired, two-tailed). (C) Scheme of V(D)J gene rearrangement and identification of shared clonotype. Shared clonotype are defined as a group of sequences with the same V/J combination event, the same heavy chain complementary determining region (HCDR3) length, and HCDR3 amino acid sequences of 80% identity between different people. (D) Lineage structure of the VH3-53-J6 clonotype. Each dot represents a unique antibody clone, two dots are connected via a line if they have 80% amino acid similarity on HCDR3 region, the dot area represents the read count for each unique antibody clone, and different colours were used to distinguish different COVID-19 donors and healthy controls. (E) The HCDR3 sequence comparison of the VH3-53-J6 clonotype in the repertoires of different COVID-19 donors. (F) Presence of the shared VH3-53-J6 clonotype in repertoires of COVID-19 patients and healthy people. (G) Identity-divergence plot of three VH3-53 antibody clones (PtL108-83, PtZ108-117, and PtK111-203) from samples collected three months after hospital discharge. All VH3-53 sequences in the repertoires were plotted as a function of sequence divergence from VH3-53 germline gene and sequence identity to PtL108-83, PtZ108-117, and PtK111-203, respectively. Colour gradient indicated sequence density. The sequence with an identical HCDR3 with PtL108-83, PtZ108-117, and PtK111-203 are shown as magenta dots.