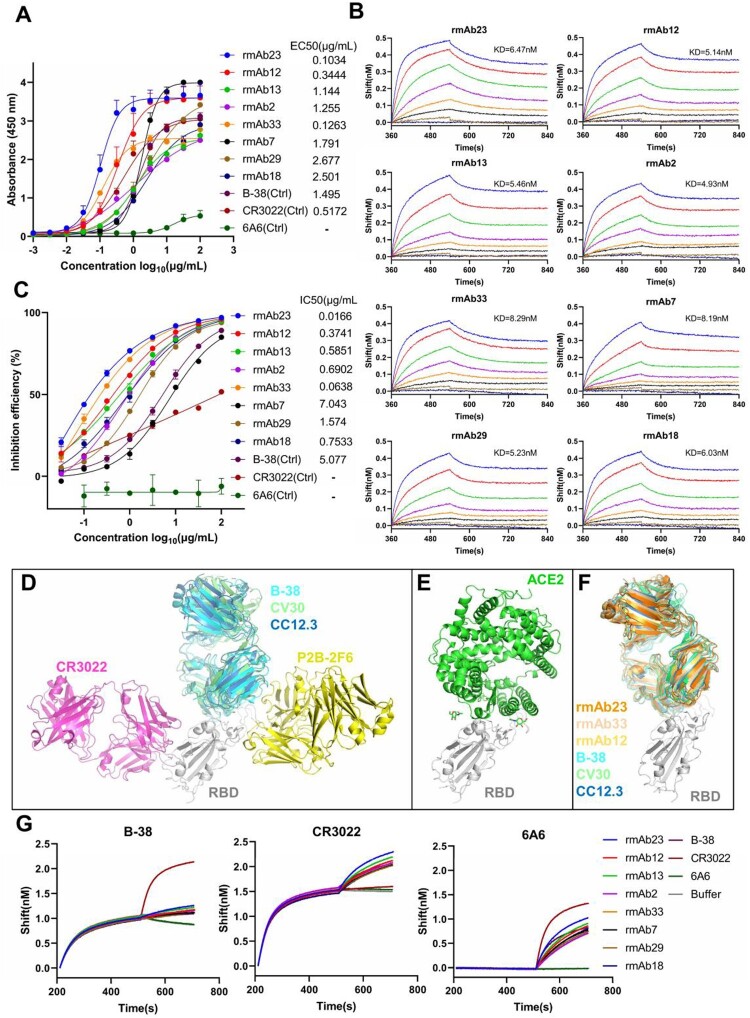
Figure 5.
VH3-53-J6 antibodies identified from the repertoires of different COVID-19 patients can bind to SARS-CoV-2 RBD and block the interaction between RBD and ACE2. (A) Eight recombinant VH3-53 antibodies with relatively higher RBD-binding activities were selected for further analysis. Previously published mAbs B-38 [11], CR3022 [33] and 6A6 [47] were used as controls. (B) Binding kinetics of the 8 recombinant VH3-53 antibodies with SARS-CoV-2 RBD were measured by BLI. Immobilized individual antibody (10 μg/mL) was saturated with RBD at seven different concentrations (200 nM, 100 nM, 50 nM, 25 nM, 12.5 nM, 6.25 nM, and 3.125 nM) or buffer. (C) Neutralizing activities of the 8 recombinant VH3-53 antibodies, determined using a SARS-CoV-2 surrogate virus neutralization test (sVNT). B-38, CR3022 and 6A6 were used as controls (D) Comparison of B-38 (PDB: 7BZ5), CV30 (PDB: 6XE1) [17], CC12.3 (PDB: 6XC4) [16], CR3022 (PDB: 6W41), and P2B-2F6 (PDB: 7BWJ) [3] binding epitopes. SARS-CoV-2 RBD is shown in cartoon representation (grey). The heavy and light chains of B-38, CV30, CC12.3, CR3022, and P2B-2F6 are coloured in cyan, limegreen, skyblue, magenta, and yellow, respectively. (E) Crystal structure of ACE2 (green) in complex with SARS-CoV-2 RBD (grey) (PDB: 6LZG) [1]. (F) Comparison of rmAb23, rmAb33, rmAb12, B-38, CV30, and CC12.3 binding epitopes. Modelling structure of rmAb23, rmAb33, and rmAb12 in complex with SARS-CoV-2 RBD are coloured in orange, wheat, yellow-orange and grey, respectively. (G) Competition binding to RBD between the 8 recombinant VH3-53 antibodies and B-38, CR3022, or 6A6 was measured by BLI, respectively. Immobilized RBD (2 μg/mL) was saturated with 10 μg/mL of the first antibody and then flowed with equal concentration of the first antibody in the presence of or without the second antibody. The graphs show binding patterns after saturation of RBD.