ABSTRACT
The newly emerging variants of SARS-CoV-2 from South Africa (B.1.351/501Y.V2) and Brazil (P.1/501Y.V3) have led to a higher infection rate and reinfection of COVID-19 patients. We found that the mutations K417N, E484K, and N501Y within the receptor-binding domains (RBDs) of the virus could confer ~2-fold higher binding affinity to the human receptor, angiotensin converting enzyme 2 (ACE2), compared to the wildtype RBD. The mutated version of RBD also completely abolishes the binding of bamlanivimab, a therapeutic antibody, in vitro. Detailed analysis shows that the ~10-fold gain of binding affinity between ACE2 and Y501-RBD, which also exits in the high contagious variant B.1.1.7/501Y.V1 from the United Kingdom, is compromised by additional introduction of the K417/N/T mutation. Mutation of E484K leads to the loss of bamlanivimab binding to RBD, although this mutation does not affect the binding between RBD and ACE2.
KEYWORDS: SARS-COV-2, COVID-19, B.1.1.7/501Y.V1, B.1.351/501Y.V2, P.1/501Y.V3, ACE2, RBD; Bamlanivimab
Introduction
The SARS-CoV-2 virus has infected over one hundred million people (COVID-19 patients) and caused more than two million deaths to date.1 The number of affected people continues to grow rapidly, emphasizing the need for rapid use of effective vaccines. Although two Spike mRNA-based vaccines (Pfizer-BioNTech and MODERNA COVID-19 vaccines) have been granted emergency use authorization in the United States,2,3 the increasing number of Spike variants that have appeared around the world has raised concerns about the continued efficacy of the vaccines.4 It has been reported that ~90% of broadly neutralizing anti-SARS-CoV-2 antibodies from COVID-19 patients engage the receptor-binding domain (RBD) of the virus Spike protein.5 Monoclonal antibodies specifically targeting the native form of the Spike developed by Regeneron and Eli Lilly have been authorized for emergency use by the Food and Drug Administration.6–9
An N501Y variant of SARS-CoV-2 (B.1.1.7, 20I/501Y.V1) that first emerged in the United Kingdom is now spreading to the rest of the world, and it appears to be much more contagious than the original virus.4 We found that this N501Y single mutation confers an ~10-fold increase in affinity between RBD and ACE2.10 However, this mutation does not affect its binding to the therapeutic antibody bamlanivimab.10 We concluded that the increase in high binding affinity may account for the high infection rate of the United Kingdom variant, while both vaccines and bamlanivimab should retain their ability to combat this newly emerging variant.10 However, this same N501Y mutation is also found in a variant (B.1.351, 20 H/501Y.V2) with mutations of K417N, E484K, and N501Y from South Africa and a variant (P1, 20 J/501Y.V3) with K417T, E484K, and N501Y from Brazil with additional mutations within the RBD.4 The two additional mutations, K417N/T and E484K, are also critical residues involved in the interactions between RBD and ACE2, as well as bamlanivimab. It has been reported that a COVID-19 patient was infected a second time by the new variant from Brazil.11 This information raises substantial concerns as to whether current vaccines and therapeutic antibodies will remain effective. Here, we address the basis of how these two additional mutations at K417 and E484 from both the South African and Brazilian variants affect the binding of RBD to ACE2 and bamlanivimab.
Results
In February 2021, we reported that N501Y-RBD (N501 mutated to Y501) derived from the United Kingdom variant has an ~10-fold increased binding affinity (0.566 nM) toward ACE2 compared to the wildtype (5.768 nM).10 Another early report based on deep mutational scanning of RBD of SARS-COV-2 also found that the mutation of N501 to Y/F501 enhances the binding of RBD to ACE2, suggesting that aromatic rings of both Y and F could account for the increase in binding affinity,12 consistent with our analysis.10 It was reported that both the South African variant and the Brazilian variant are also as highly contagious as the United Kingdom variant.13
In this study, our goal was to determine whether these two additional mutations within the RBD region affect the binding affinity between RBD and ACE2 and could account for the high infection rate. On the basis of Y501-RBD, two additional mutations, K417N and E484K existing in the South African variant, were introduced and expressed in 293 F cells as previously reported.10 Purified protein N417/K484/Y501-RBD was subjected to binding assays to ACE2 on a Biacore machine. The binding affinity (2.935 nM) between N417/K484/Y501-RBD and ACE2 is much lower than that of Y501-RBD and ACE2 (0.566 nM), although still ~2-fold higher than that between wildtype RBD (N501-RBD) and ACE2 (5.768 nM) (Figure 1(a)). Nevertheless, this ~2-fold binding affinity increase may partially account for the higher infectious rate presenting in South Africa and Brazil.
Figure 1.
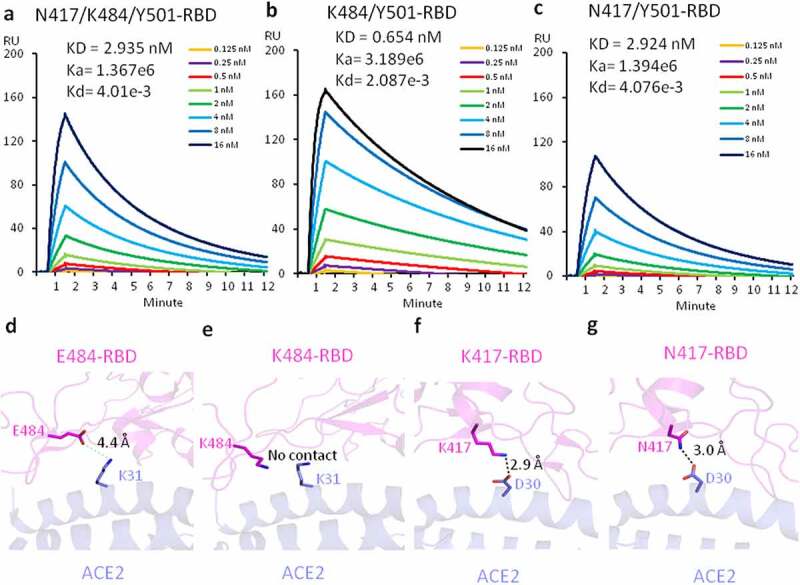
The affinity measurement and structural modeling between the mutated RBDs and ACE2. (a) The Binding of N417/K484/Y501-RBD and ACE2. (b) The binding of K484/Y501-RBD and ACE2. (c) The binding of N417/Y501-RBD and ACE2. (d) The model of interactions between E484-RBD and ACE2. (e) The model of interactions between K484-RBD and ACE2. (f) The model of interactions between K417-RBD and ACE2. (g) The model of interactions between N417-RBD and ACE2
Understanding how these two additional mutations weaken the association between RBD and ACE2 is of great interest. To investigate the contribution of each additional mutation to the binding affinity, we carried out the following experiments. First, we introduced only the E484K mutation into the existing N501Y-RBD to generate a double mutation of RBD (484 K/501Y-RBD), which exist in both the South African and Brazilian variants, and expressed it in 293 F cells. Again, purified protein was subjected to surface plasmon resonance (SPR) binding assays to examine binding affinity between 484 K/501Y-RBD and ACE2. As this E484K mutation incorporates the positively charged residue lysine in the place of the negatively charged residue glutamic acid, we expected a drastic drop of binding affinity between 484 K/501Y-RBD and ACE2. To our surprise, the binding affinity between this double mutation version of RBD (K484/Y501-RBD) and ACE2 is 0.654 nM (Figure 1(b)), an affinity that is similar to that of Y501-RBD and ACE2 at 0.566 nM, as we previously reported.10 This result suggests the mutation does not affect the binding between RBD and ACE2.
To verify this conclusion, we explored the detailed interactions of K484/Y501-RBD and ACE2 and carried out molecular docking based on the published complex structure of RBD and ACE2.14 As shown in Figure 1(d), there could be a weak salt bridge (4.4 Å, longer than 4.0 Å) formed between E484 of RBD and K31 from ACE2. After conversion of E484 to K484, this weak salt bridge is gone (Figure 1(e)), although this interaction is too weak to have obvious impact on the receptor binding. We then proceeded to introduce another mutation, K417N within the South African variant, on the basis of 501Y-RBD. As before, a double mutation version with K417N and N501Y protein (N417/Y501-RBD) was produced and binding to ACE2 was assessed by SPR. Confirming our expectation, this additional mutation of K to N drastically reduced the binding affinity (2.924 nM) between N417/Y501-RBD and ACE2 compared to 0.566 nM for the Y501-RBD to ACE2 interaction (Figure 1(c)). Again, we did molecular docking analysis of this mutation. The original salt bridge formed between K417 of RBD and D30 of ACE2 was lost in the new interaction, but was replaced by a new weaker hydrogen bond (Figure 1(f) and (g)).
Overall, the changes of affinity with each mutation could be interpreted by the structural modeling data. We conclude that the three mutations of RBD from the South African variant lead to an ~2-fold increase of binding affinity between the original RBD and ACE2, which may account for the higher infection rate of the South African variant over the original virus. However, the affinity to ACE2 of this new variant is decreased by a factor of fivefold compared to the United Kingdom variant.10 We demonstrated here how each individual muation contributes to the change of ACE2 binding affinity either by enhancing or reducing it.
Several monoclonal antibodies targeting SARS-CoV-2 have been granted emergency authorization as treatments for COVID-19 patients.6–9 Most importantly, all these therapeutic antibodies are RBD specific. We investigated the binding of the mutant RBD with the therapeutic antibodies to verify the efficacy of these antibodies. The binding assays between three versions of RBD proteins with different mutation combination and bamlanivimab were generated by applying them to immobilized antibodies on the Biacore Chip (Figure 2). To our surprise, the interaction between the mutant RBD containing three mutations, N417/K484/Y501-RBD, and bamlanivimab, was completely abolished (Figure 2(a)). However, the binding between N417/Y501-RBD and antibody remains the same as those between wildtype N501-RBD or mutated Y501-RBD and the antibody with binding affinities of 0.976 nM (Figure 2(b)), 0.874 nM and 0.801 nM, respectively.10 Furthermore, when the E484K mutation is introduced, the binding between K484/Y501-RBD and the antibody is completely abolished (Figure 2(a) and (c)).
Figure 2.
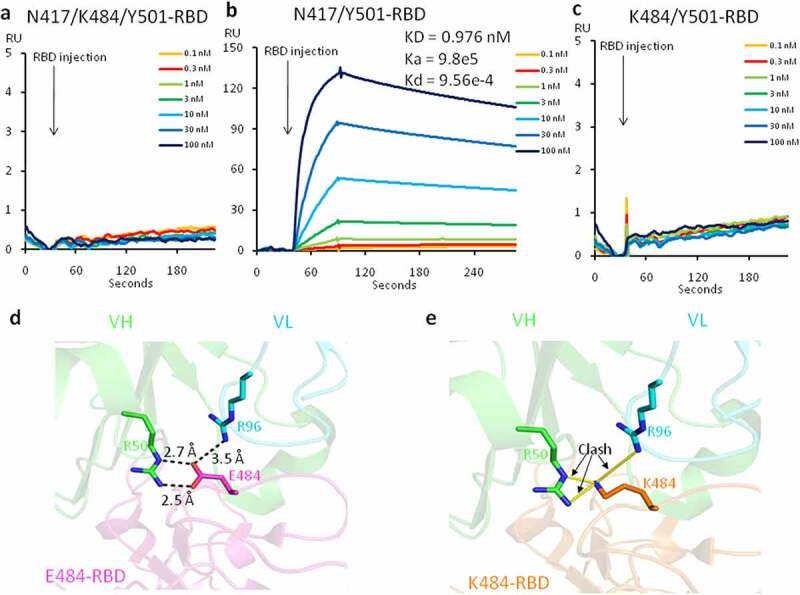
The affinity measurement and structural modeling between the mutated RBDs and the therapeutic antibody, bamlanivimab. (a) The binding between N417/K484/Y501-RBD and bamlanivimab. (b) The binding between N417/Y501-RBD and bamlanivimab. (c) The binding between K484/Y501-RBD and bamlanivimab. (d) The model of interactions between E484-RBD and bamlanivimab. (e) The model of interactions between K484-RBD and bamlanivimab. VH, variable heavy chain. VL, variable light chain
The structure of the entire Spike protein and bamlanivimab has been reported.15 We used the available complex structure to analyze how this critical E484K mutation causes a loss of binding. The structure showed that E484 of RBD forms three salt bridges with variable heavy chain R50 (VH R50) and variable light-chain R96 (VL R96) of bamlanivimab (LY-CoV555) (Figure 2(d)). Our prediction shows that the conversion of E484 of RBD to K484 leads to the loss of three salt bridges, as well as the formation of a new repulsive charge–charge interaction brought in from both positively charged sidechains of Lys and Arg (Figure 2(e)). This easily can explain the dramatic drop or loss of binding between RBD and the therapeutic antibody. Our binding data and structural analysis suggests that bamlanivimab (LY-CoV555) completely loses its efficacy in COVID-19 patients infected with the South African and Brazilian variants.
Discussion
During 2020 and 2021, the COVID-19 pandemic has had disastrous effects worldwide, and there are still limited means to contain it. The mutation rate of the virus is low, but mutations in critical sites still could threaten the efficacy of vaccines and therapeutic neutralizing antibodies. We found that a single mutation, N501Y, found in the United Kingdom variant, conferred an ~10-fold higher binding affinity of the RBD of the SARS-CoV-2 Spike protein to ACE2, the receptor from human host, compared to that of wildtype RBD and ACE2.10 This dramatic increase of binding affinity could account for the much higher infection rate of this variant. Interestingly, this mutation was also found in both the South African variant (B.1.351/501Y.V2) and the Brazilian variant (P1/501Y.V3), and therefore, it may partially explain why these two variants are also more contagious than the wildtype.13 To our surprise, the additional K417N mutation within RBD region of the Spike from the South African variant lowers the binding affinity of this RBD (417 N/484 K/501Y-RBD) to ACE2 almost fivefold, although it remains ~ 2-fold higher than that of wildtype to ACE2. Due the high similarity between the South African variant (417 N/484 K/501Y-RBD) and the Brazilian variant (417 T/484 K/501Y-RBD), we believe that the latter should have similar binding affinity to ACE2 as that of the South African variant, loss of salt bridge and consequently decrease in the interaction between RBD and ACE2. Interestingly, detailed analysis showed that introduction of the E484K mutation alone within RBD (K484/Y5010-RBD) does not affect the binding affinity to ACE2, while a single mutation of K417N in the South African variant confers a dramatic drop of the binding affinity. The introduction of K417T existing from the Brazilian variant should lead to the similar drop due to the loss of potential salt bridges.
On the other hand, although the E484K mutation does not affect the binding between RBD and ACE2, it completely abolishes the binding of RBD to bamlanivimab. This in vitro data may indicate that this antibody will not effectively treat COVID-19 patients with either the South African variant or the Brazilian variant. Another alarming signal is that the area around E484 on RBD is a hot spot for broadly neutralizing antibodies, and several other authorized therapeutic antibodies also target this region (data not shown). This may also explain why the Brazilian variant could re-infect COVID-19 patients.11
Many researchers are currently focused to the efficacy of broadly neutralizing antibodies generated by COVID-19 patients and vaccines. As demonstrated by Sette and colleagues,16 the T cell arm of adaptive immunity plays a similar critical, if not the more important role, in combatting the virus. While B cells use strictly 3-dimentional (3D) epitopes from the virus, helper T cells and cytotoxic T cells use linear epitopes in their fight against the virus. Thus, T cells, with their completely different protection mechanism through conserved linear antigens derived from SARS-CoV-2, could be effective against newly emerging and slowly evolved SARS-CoV-2 variants, making the loss of efficacy of antibodies a less frightening prospect. In this regard, vaccines that provide a large group of T cell antigens of the entire virus, rather than just the Spike protein, e.g., inactivated virus vaccines, should not be forgotten. Overall, we believe that broad vaccinations with mRNA-based vaccines or others will build-up herd immunity to beat SARS-CoV-2.
This study shows the danger of mutations within RBD from the aspect of efficacy of therapeutic antibodies. These therapeutic antibodies were derived from a few conserved 3D epitopes of the virus, mostly from the RBD domain. We showed previously that higher concentrations of the antibody may be needed to treat COVID-19 patients infected with the United Kingdom variant compared to patients with wildtype virus, even though N501Y mutation does not affect the binding of bamlanivimab.10 However, these additional mutations within the RBD regions, such as E484K besides N501Y within both the South African and Brazilian variants, cause complete loss of efficacy of bamlanivimab. We acknowledge that our in vitro data requires further verification in vivo. Besides bamlanivimab, E484K mutation has been reported to escape the recognition of more monoclonal antibodies targeting RBD region.17 The extent of escape should correlate with how much E484 contributes to the RBD and antibody interaction, as E484K mutation would not only lose the negatively charged glutamic acid side chain, but add positively charged lysine side chain. From our perspective, therapeutic antibodies to treat COVID-19 patients should be adjusted for the new emerging variants accordingly.
Materials and Methods
SARS-CoV-2 spike RBD mutation and expression
SARS-CoV-2 RBD (319–541aa) was cloned to pCDNA3.1 vector with a 6-histidine tag at the C-terminal. Y501 mutation was created by quick change mutagenesis and verified by DNA sequencing. N417 or K484 was mutated on the Y501-RBD backbone by the same method to create RBD with double mutation sites. The triple mutation RBD was created in the same manner. All the mutations were verified by DNA sequencing. The mutated RBD was transiently expressed in 293 F cell line by transfection. The supernatant was used for RBD purification by passing through a nickel column. The eluted RBD was further purified by Superdex-200 Gel-filtration size column.
Biacore affinity measurement of mutant RBD binding to ACE2
Affinity measurements were carried out with Biacore T200. Biotinylated BirA tag-ACE2 (1–615) was used to coat the Strepavidin chip.10 Mutant RBD was injected at different concentrations. The affinity was calculated by the BIA evaluation software.
Biacore affinity measurement of mutant RBD binding to bamlanivimab
Affinity measurement was carried out with Biacore T200. Bamlanivimab was coated on the CM5 chip by the amine coupling method.10 Mutant RBD was injected at different concentrations. The bound RBD was eluted with 10 mM glycine pH 1.7. The affinity was calculated by the BIA evaluation software.
Protein docking and binding affinity prediction for mutants
Protein docking studies were performed by HADDock 2.2 server.18 The crystal structure of SARS-CoV-2 RBD bound with ACE2 (PDB code: 6M0J) was selected for docking. Two different mutant (N501Y_K417N and N501Y_E484) structures were prepared by Coot software.19 One of the conformations from alternative residues in the structure was deleted manually to meet the docking server criteria. The major contact residues in 6M0J (hydrogen bond and salt bridge) were selected as restrain residues for docking (Q24, E35, E37, D38, Y41, Q42 and Y83 for ACE2 protein; N487, Q493, Y449, and Y505 for RBD protein). The best model from the docking was selected for the next analysis. All protein structural figure prepared by PyMOL. The binding affinity prediction for each of mutants was did by BeAtMuSiC server.20
Acknowledgments
We thank National Jewish Health for support and people in the Kappler/Marrack groups for help. H.L. is partially supported by NIH grant (GM135421 to G.Z,) and NB Life Laboratory LLC. We also thank the Colorado Department of Public Health and Environment (CDPHE) for authorization to use the residual therapeutic antibodies including bamlanivimab after COVID-19 patient infusions.
Funding Statement
This work was supported by the National Institute of General Medical Sciences [GM135421].
Contributions
H.L., P.M., and G.Z. for designing; H.L. for main experiments; P.W., Q.Z., Z.C., K.A., W.D., S.P., G.D., L.R., S.F. for some experiments; H.L., P.M., G.Z. for final data analysis and writing up.
Disclosure of Potential Conflicts of Interest
H.L. is partially supported by NB Life Laboratory LLC, G.Z. holds equity at NB Life Laboratory LLC. We do not have any financial relation with Pfizer-BioNTech, Moderna, Eli Lilly, or Regeneron.
References
- 1.Worldometer . COVID-19 Coronavirus Pandemic. Accessed 2021 April10. https://www.worldometers.info/coronavirus/.
- 2.Polack P, Thomas S, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez J, Marc G, Moreira E, Zerbini C, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med. 2020;383(27):2603–6. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baden L, Sahly H, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector S, Rouphael N, Creech C, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403–16. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention . Science brief: emerging SARS-CoV-2 variants. Accessed 2021 April10. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html. [PubMed]
- 5.Piccoli L, Park Y, Tortorici M, Czudnochowski N, Walls A, Beltramello M, Fregni C, Pinto D, Rosen L, Bowen J, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4):1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum A, Ajithdoss D, Copin R, Zhou A, Lanza K, Negron N, Ni M, Wei Y, Mohammadi K, Musser B, et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370(6520):1110–15. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb R, Nirula A, Chen P, Boscia J, Heller B, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325(7):632. doi: 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.US Food and Drug Administration . Bamlanivimab re-issued authorization letter dated 2021. March 2. Accessed 2021 April10. https://www.fda.gov/media/143602/download.
- 9.US Food and Drug Administration . Coronavirus (COVID-19) update: FDA authorizes monoclonal antibodies for treatment of COVID-19. 2020 November 21, press release. Accessed 2021 April10. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 (2020).
- 10.Liu H, Zhang Q, Wei P, Chen Z, Aviszus K, Yang J, Downing W, Peterson S, Jiang C, Liang B, et al. The basis of a more contagious 501Y.V1 variant of SARS-COV-2. Cell Res (2021). 10.1038/s41422-021-00496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Resende P, Bezerra J, Vasconcelos T, Arantes I, Appolinario L, Mendonca A, Paixao A, Rodrigues A, Silva T, Rocha A, et al. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020. Accessed 2021 April10. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584.
- 12.Starr T, Greaney A, Hilton S, Ellis D, Crawford K, Dingens A, Navarro M, Bowen J, Tortorici M, Walls A, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310 e1220. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San E, Msomi N, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv. 2020. doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 14.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–20. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 15.Jones B, Brown-Augsburger P, Corbett K, Westendorf K, Davies J, Cujec T, Wiethoff C, Blackbourne J, Heinz B, Foster D, et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv. 2020. doi: 10.1101/2020.09.30.318972. [DOI] [Google Scholar]
- 16.Grifoni A, Weiskopf D, Ramirez S, Mateus J, Dan J, Moderbacher C, Rawlings S, Sutherland A, Premkumar L, Jadi R, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501 e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H, Zhang Y, Li T, Liu S, Zhang M, et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:1–10. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zundert G, Rodrigues J, Trellet M, Schmitz C, Kastritis P, Karaca E, Melquiond A, Dijk M, Vries S, Bonvin A.. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428(4):720–25. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Emsley P, Cowtan K. Coot : model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 20.Dehouck Y, Kwasigroch J, Rooman M, Gilis D. BeAtMuSiC: prediction of changes in protein–protein binding affinity on mutations. Nucleic Acids Res. 2013;41(W1):W333–339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]


