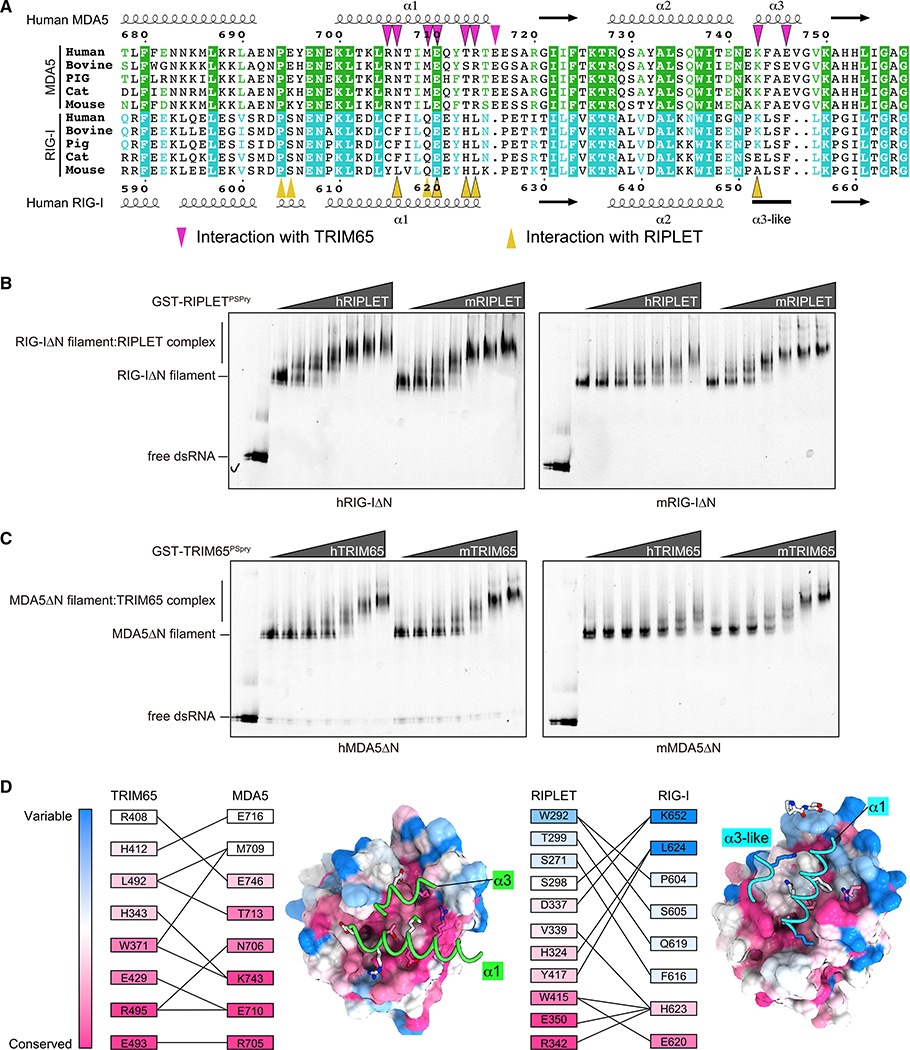
Figure 4. Conservation and co-evolution analysis of the RLR:PSpry interface.
(A) Sequence alignment of orthologs of MDA5 and RIG-I near the TRIM65/RIPLET interface. Residues involved in the interaction with TRIM65 and RIPLET are indicated with triangles Sequences were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo), and alignment figures were generated using ESPript3 (http://espript.ibcp.fr/ESPript/ESPript).
(B) Native gel mobility shift assays of RIG-I filaments in the presence of RIPLET. RIG-I filaments were assembled by incubating Cy5-labeled 112-bp dsRNA (1 ng/μL) with human or mouse RIG-IΔN (hRIG-IΔN or mRIG-IΔN, respectively; 250 nM) in the presence of 2 mM ATP (Peisley et al., 2013). RIG-I filaments were then incubated with increasing concentrations (18–600 nM) of human or mouse RIPLETPSpry (hRIPLET or mRIPLET, respectively) fused with GST, and complex formation was analyzed by native PAGE using dsRNA fluorescence.
(C) Native gel mobility shift assay of MDA5 filaments in the presence of TRIM65. Experiments were done as in (A) except that ATP was omitted in the reaction because ATP promotes MDA5 filament disassembly (Peisley et al., 2011).
(D) Degree of conservation for interacting residues in the MDA5:TRIM65 and RIG-I:RIPLET complexes. The conservation score was calculated using vertebrate protein sequences (STAR methods) and the Consurf web server (https://consurf.tau.ac.il/). The interacting residues are arranged in the descending order of conservation score (bottom to top), and the interaction pairs are indicated by connecting lines. The conservation score was also mapped onto the structures of PSpry using the program CueMol. The PSpry domains of TRIM65/RIPLET are shown in surface representation from the equivalent viewpoints, whereas α1/α3 helices of the bound RLRs are shown in cartoon representation.