Fig. 2.
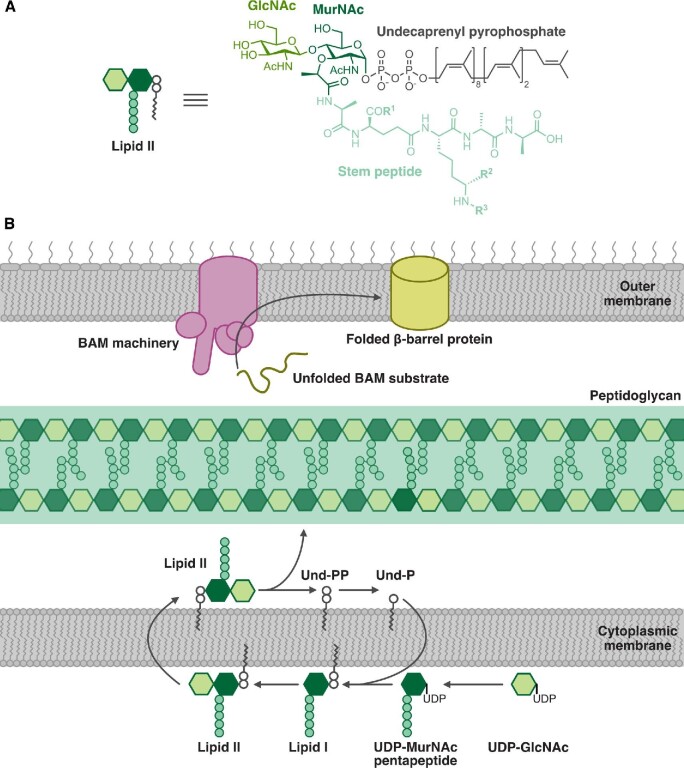
The bacterial cell envelope is a common target for RiPPs. (A) Cartoon and chemical structure of lipid II, the precursor to peptidoglycan. The stem peptides of lipid II from different bacteria vary at the positions indicated by R1, R2, and R3. GlcNAc: N-acetylglucosamine, MurNAc: N-acetylmuramic acid. (B) Schematic of cell envelope biosynthesis processes disrupted by RiPPs. Both lanthipeptides and the lasso peptide siamycin-I bind to lipid II, disrupting peptidoglycan biosynthesis. The recently discovered RiPP darobactin inhibits BamA, disrupting the assembly of outer membrane proteins in Gram-negative bacteria. BAM: β-barrel assembly machinery.