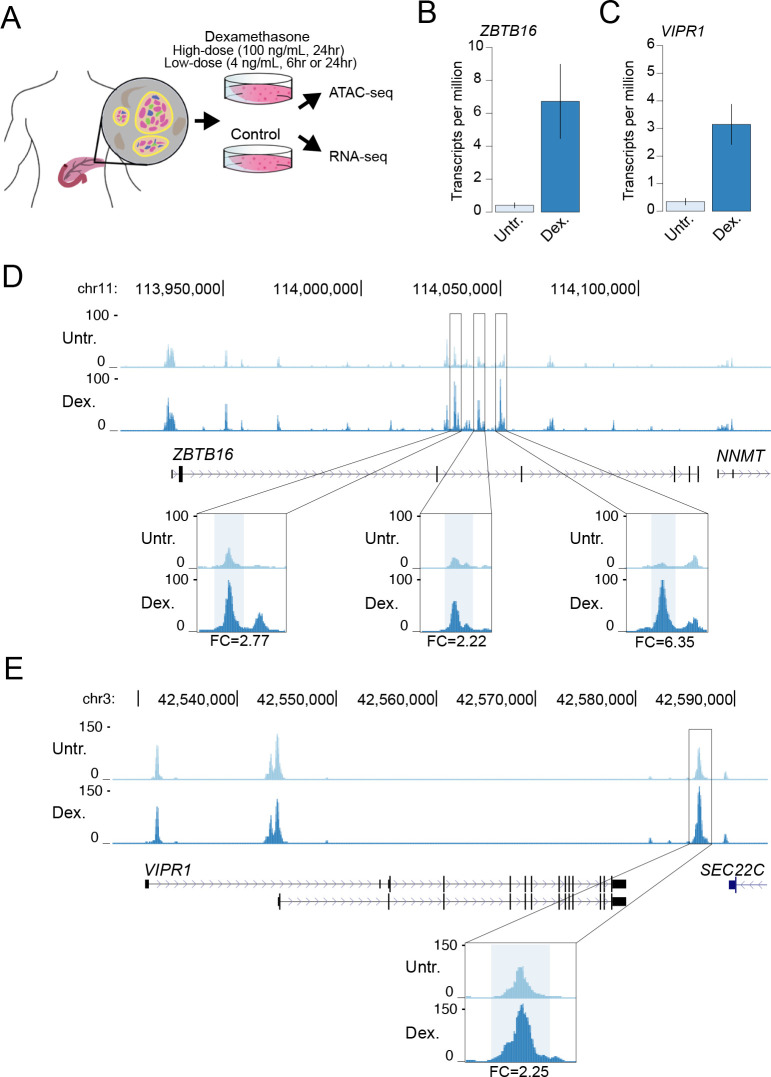
Fig 1. A map of gene regulation in pancreatic islets in response to glucocorticoid signaling.
(A) Overview of study design. Primary pancreatic islet samples were split and separately cultured in normal conditions and including the glucocorticoid dexamethasone at either a high-dose (100ng/mL for 24hr) or low-dose (4 ng/mL for 6hr or 24hr) treatment, and then profiled for gene expression and accessible chromatin using RNA-seq and ATAC-seq assays. Genes with known induction in glucocorticoid signaling (B) ZBTB16 and (C) VIPR1 had increased expression in glucocorticoid-treated islets compared to untreated islets. Values represent mean and standard error. (D) At the ZBTB16 locus several accessible chromatin sites intronic to ZBTB16 had increased accessibility in glucocorticoid treated (Dex.) compared to untreated (Untr.) islets. (E) At the VIPR1 locus an accessible chromatin site downstream of VIPR1 had increased accessibility in glucocorticoid treated (Dex.) compared to untreated (Untr.) islets. Values in D and E represent RPKM normalized ATAC-seq read counts. Fold-change (FC) in accessible chromatin signal in glucocorticoid treatment compared to untreated indicated at highlighted sites. All results shown are for the high-dose treatment.