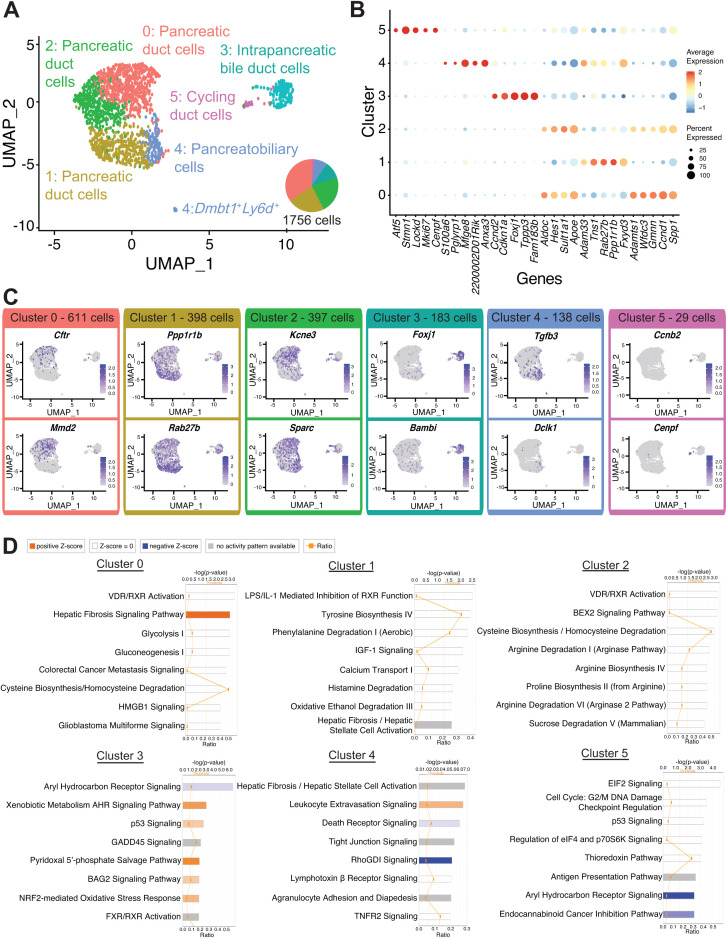
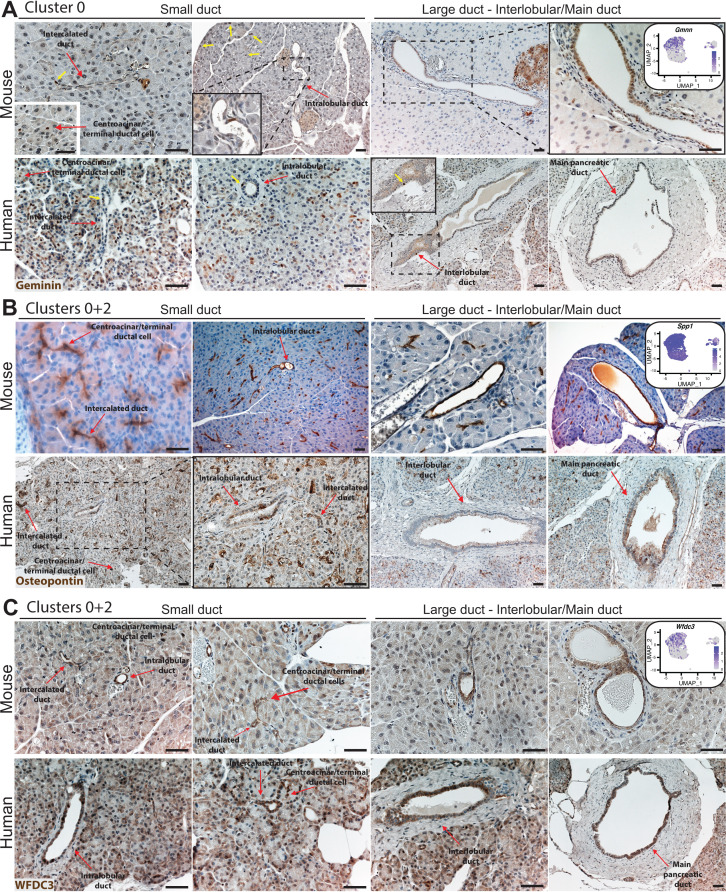
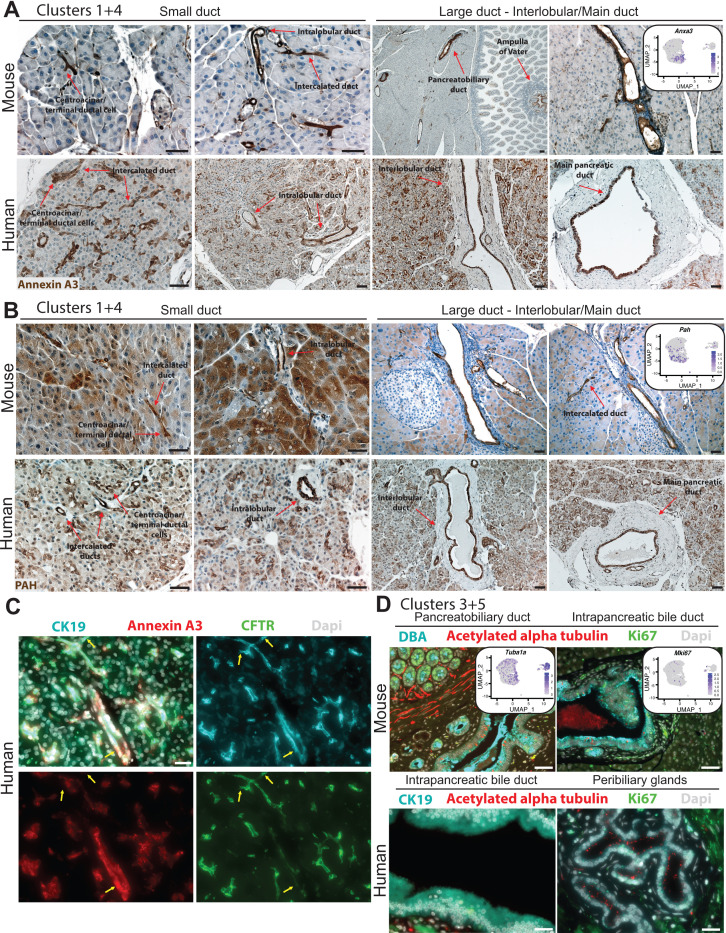
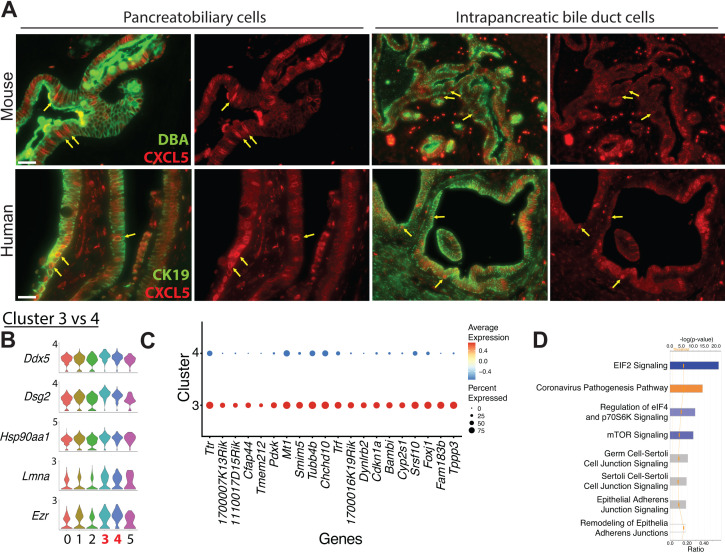
Figure 2. Transcriptomic map of DBA+pancreatic duct cells.
(A) Uniform Manifold Approximation and Projection (UMAP) depicts identity of clusters. (B) The dot plot shows the top five significantly differentially expressed genes (DEGs) with the highest fold change for each cluster. (C) Feature plots show expression of significantly DEGs for clusters 0, 1, 3, 4, and 5. Cluster 2 is characterized by lack of or low-level expression of significantly DEGs found in other clusters. (D) Ingenuity pathways analysis (IPA) results show the top eight deregulated pathways when comparing a cluster to all other clusters. The ratio line indicates the fraction of molecules significantly altered out of all molecules that map to the canonical pathway from within the IPA database. A positive z-score represents upregulation, and a negative z-score indicates downregulation of a pathway in that cluster when compared to all other clusters. A gray bar depicts significant overrepresentation of a pathway, the direction of which cannot yet be determined.