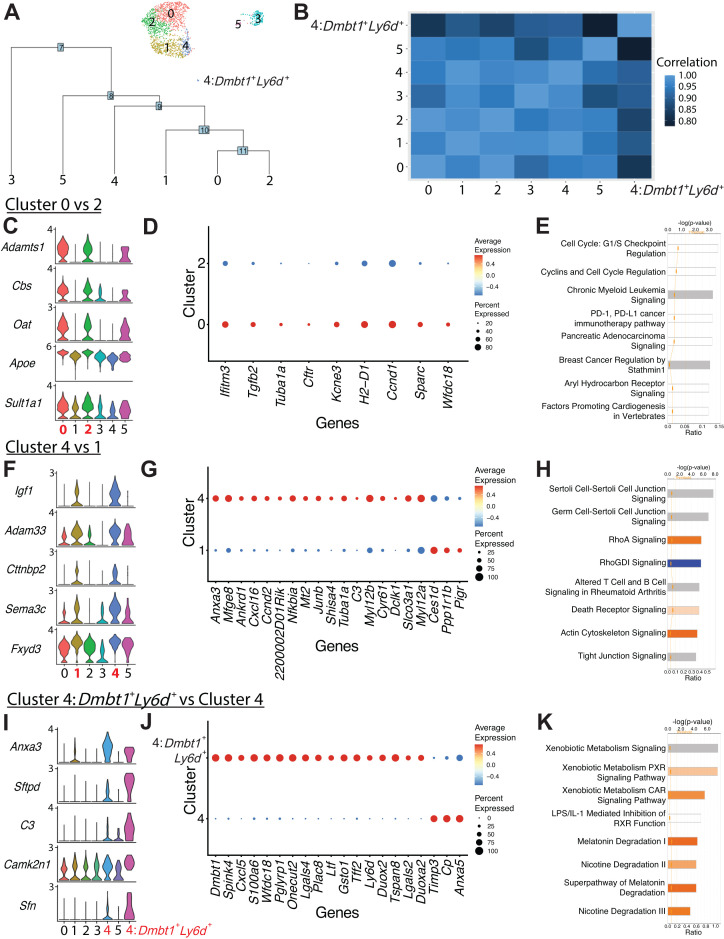
Figure 3. Comparison of ductal clusters 0 vs. 2, 4 vs. 1, and 4 vs. 4:Dmbt1+Ly6d+.
(A) The cluster dendrogram created using dims (used to define the cluster) shows the Euclidean relationships between clusters. The tree is calculated in the principal component analysis space. The genes used to define the tree were set as the variable features of the object. (B) Pearson’s correlation calculated using average gene expression is depicted. (C) Stacked violin plots show five differentially expressed genes (DEGs) sharing similar expression patterns in clusters 0 and 2. (D) The dot plot shows all nine DEGs found when comparing cluster 0 vs. 2. (E) The top eight altered pathways from ingenuity pathways analysis (IPA) comparing cluster 0 vs. 2 are depicted. (F) Stacked violin plots show five DEGs sharing similar expression patterns in clusters 4 and 1. (G) The dot plot shows the top 20 DEGs ranked by fold change when comparing cluster 4 vs. 1. (H) The top eight deregulated pathways from IPA comparing cluster 4 vs. 1 are depicted. (I) Stacked violin plots of five DEGs sharing similar expression patterns in clusters 4:Dmbt1+Ly6d+ and 4. (J) The dot plot shows the top 20 DEGs ranked by fold change when comparing clusters 4:Dmbt1+Ly6d+ and 4. (K) The top eight changed pathways from IPA comparing clusters 4:Dmbt1+Ly6d+ and 4 are depicted.