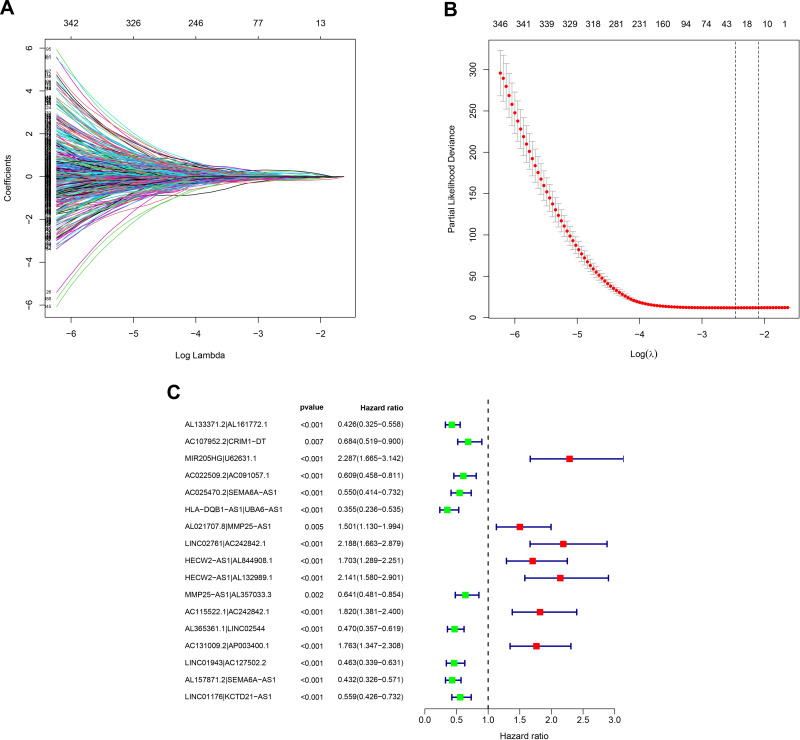
Figure 2.
Identification of survival-related IRlncRs using LASSO regression analysis and multivariate Cox regression analysis. (A) LASSO coefficient profiles of the 28 sIRlncRs of SKCM. (B) Plots of the cross-validation error rates. Each dot represents a lambda value along with error bars that represent the confidence interval for the cross-validated error rate. The top of the plot gives the size of each model. The vertical dotted line indicates the value with the minimum error and the largest lambda value where the deviance is within one SE of the minimum. (C) Forest plots of HR of sIRlncRs pairs obtained by multivariate Cox regression analysis. A total of 6 DEGs were found to be prognostic factors. The genes with HR < 1 are protective factors, while the ones with HR > 1 are risk factors in SKCM.
Abbreviations: IRlncRs, immune-related long non-coding RNAs; LASSO, least absolute shrinkage and selection operator; SKCM, skin cutaneous melanoma; HR, hazard radio; DEGs, differentially expressed genes.