Abstract
Background:
Many microRNAs (miRNAs) have been associated with asthma and chronic obstructive pulmonary disease (COPD). Longitudinal lung function growth trajectories of children with asthma—normal growth, reduced growth (RG), early decline (ED), and RG with an ED (RGED)—have been observed, with RG and RGED associated with adverse outcomes, including COPD.
Objective:
Our aim was to determine whether circulating miRNAs from an early age in children with asthma would be prognostic of reduced lung function growth patterns over the next 16 years.
Methods:
We performed small RNA sequencing on sera from 492 children aged 5 to 12 years with mild-to-moderate asthma from the CAMP clinical trial, who were subsequently followed for 12 to 16 years. miRNAs were assessed for differential expression between previously assigned lung function growth patterns.
Results:
We had 448 samples and 259 miRNAs for differential analysis. In a comparison of the normal and the most severe group (ie, normal growth compared with RGED), we found 1 strongly dysregulated miRNA, hsa-miR-145-5p (P < 8.01E–05). This miR was downregulated in both ED groups (ie, ED and RGED). We verified that miR-145-5p was strongly associated with airway smooth muscle cell growth in vitro.
Conclusion:
Our results showed that miR-145-5p is associated with the ED patterns of lung function growth leading to COPD in children with asthma and additionally increases airway smooth muscle cell proliferation. This represents a significant extension of our understanding of the role of miR-145-5p in COPD and suggests that reduced expression of miR-145-5p is a risk factor for ED of long-term lung function.
Keywords: Asthma, COPD, miRNA, lung growth patterns, Childhood Asthma Management Program, airway smooth muscle
GRAPHICAL ABSTRACT
MicroRNAs (miRNAs) are small noncoding RNAs that regulate their target mRNAs posttranscriptionally through degradation or translational repression.1 miRNAs have emerged as vital molecules in asthma and chronic obstructive pulmonary disease (COPD).2 Approximately 60% of mRNAs may be the targets of miRNAs. They have a potential role in regulating the signaling pathways in different types of cells, including cells of the immune system, as well as in controlling the inflammatory response in various tissues.3,4
Determining the FEV1 value is a way of measuring lung function: in normal persons, it is characterized by rapid growth in childhood and adolescence, with a leveling off or plateauing in early adulthood and a gradual decline into middle and old age. In individuals with lung disease, including asthma, deviations from the canonic normal growth (NG) pattern can appear as reduced growth (RG), early decline (ED), or a combination of RG and ED. A pattern of FEV1 value increase and decline characterized by ED has been linked with smoking and respiratory symptoms, and reduced childhood lung function has been associated with an increased incidence of later-life COPD.5 Even apart from the association with later COPD, a reduced FEV1 value is clinically significant, occurring with increased asthma symptoms and exacerbations, as well as with increased mortality.6 Reduced lung function is associated with asthma incidence, asthma recurrence, and recurrent wheeze.7–11
We have previously characterized the serial lung function trajectories of children with asthma over the course of 16 years, from childhood into early adulthood, into 4 patterns: NG, RG, ED, and RG with an ED (RGED). We have shown that RG patterns can lead to fixed obstruction consistent with COPD before the age of 30 years. The purpose of this study was to determine whether circulating miRNAs from serum12 taken from children with asthma at an early age would be prognostic of those children at greatest risk of chronically low lung function.
METHODS
CAMP
The Childhood Asthma Management Program (CAMP) was a randomized, placebo-controlled trial of inhaled anti-inflammatory treatments for mild-to-moderate childhood asthma followed by 3 phases of observational follow-up; the trial and all follow-up phases included at least annual spirometry. A total of 1,041 participants enrolled in the trial between 1993 and 1995 (when they were 5 to 12 years old); follow-up continued to 2012, when the participants were 22 to 30 years old.13 We previously categorized CAMP participants into 4 patterns of lung function growth and decline based on prebronchodilator FEV1 value over the course of observation: NG, NG with an ED, RG, and RG with an ED (RGED). NG was defined as an FEV1 growth curve predominantly above the 25th percentile of the normal FEV1 value, whereas RG was defined as an FEV1 growth curve below the 25th percentile. The presence of ED was indicated by a decrease from the maximal level earlier than expected and before age 23. For further details, see McGeachie et al.9,10
To identify subjects in CAMP with spirometry consistent with COPD at the end of observation, we considered the FEV1 and forced vital capacity (FVC) values of participants who were at least 23 years of age at their final visit, following previous work.9,10 We used a robust locally weighted scatterplot, smoothing, regression method to smooth the FEV1 value and FEV1/FVC ratio for each participant across all time points.14 To obtain the most robust estimates of final-visit spirometry, we used the smoothed values at the final time point. COPD was indicated with an FEV1/FVC ratio less than the lower limit of normal, defined as the 5th percentile of FEV1/FVC ratio for a person of the same age, sex, height, and race, according to Hankinson et al,15 and an FEV1 value less than the 70% of predicted FEV1 value for a person with the same characteristics.
Data collection, QC, filtering, and normalization
We performed small RNA sequencing (RNA-seq) on baseline serum samples from 492 children from CAMP. Serum samples were stored in freezers at −80°C at the Channing Division of Network Medicine. Recent work has shown that serum miRNAs are extremely stable over many years.16,17 Small RNA-seq libraries were prepared by using the Norgen Biotek Small RNA Library Prep Kit and sequenced on the Illumina NextSeq 500 platform. The ExceRpt pipeline18 was used for quality control (QC) of the RNA-seq data. Mapped read counts less than 5 were filtered, and miRNAs with coverage of less than 50% of all subjects were removed. All samples passed QC for the number of mapped reads and total reads, indicating that satisfactory miRNA concentration was available. Using the DESeq2 R package,19 we normalized reads by relative log2 expression. We used a guided principal component analysis algorithm for identification of batch effects in our data.20 These miRNA data have previously been deposited into the Gene Expression Omnibus under the accession number GSE134897.
Statistical analysis
Our primary outcome was the pattern assignment (RG, NG, ED, and/or RGED), which was treated as a categoric variable, and pairs of patterns were compared. Differentially expressed miRs (upregulated and downregulated miRs) between lung function patterns were identified by using the DESeq2 package19 in R with a Benjamini-Hochberg false discovery rate (FDR) multiple testing correction.21 A significance threshold of 10% FDR was used. The analysis was performed without adjustment for age, sex, height, and race because these characteristics are the basis of the lung function pattern phenotypes: patterns were assigned on the basis of FEV1 value adjusted for age, sex, height, and race following Hankinson et al.15
DESeq2 was also used for checking the differential association of our top differentially expressed miRNA with COPD status. A clustered heatmap of differentially expessed miRs was computed by using the complex heatmap R package.22 Unsupervised hierarchic clustering was used to generate the heatmap, and Pearson correlation was used as the distance metric.
Predictive models of miR-145-5p and all differentially expressed miRs were obtained by using linear regression. The model parameters were fit from the entire CAMP cohort. Predictive performance was assessed by using the convex hull23 of the area under the receiver operating characteristic curve (AUC) with CIs estimated according to DeLong et al.24 Next, 5-fold cross-validation (CV) of model parameters (β-values) was performed by using MATLAB R2020a (Natick, Mass) with 100 repeats for assessing the distribution of CV performance.
miRNA target identification and functional analysis
We used Dianna micro T-CDS25,26 for putative mRNA target identification for differentially expressed miRNAs, with a 0.9 micro T-CDS (miTG) score as a threshold. We subsequently used DAVID 6.827 for functional enrichment analysis of the putative targets. For each of the Gene Ontology (GO) functional groups,28 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways,29 and InterPro functional30 terms returned from DAVID functional analysis, we considered a P value threshold of .05 or lower and a minimum gene count of 3 to be required. DAVID clusters with annotation cluster score (negative log10 P value) of at least 1.3 were considered significant. For comparison, pathway enrichment of putative targets of miR-145-5p was also investigated by using TargetScan,31 Tarbase,32 and miRWalk.33
Target genes of miR-145-5p were assessed for overlap with genes identified as being associated with pulmonary conditions from genome-wide association study (GWAS) results by using the National Human Genome Research Institute–European Bioinformatics Institute catalog (https://www.ebi.ac.uk/gwas/). Associations were assessed by using chi-square tests.
In vitro analysis
The top miRNA was then assessed in vitro for effect on human airway smooth muscle (HASM) cell proliferation and hypertrophy. HASM cells were transfected with 10 nM of either scramble control (All-Stars Negative Control siRNA, Qiagen, Venlo, The Netherlands) or miR mimic (Qiagen) by using RNAiMax (ThermoFisher Scientific, Waltham, Mass) according to the manufacturer’s protocol. This design should control for the effect of transfection on cell death. Then, 72 hours after transfection, the cells were trypsinized and measured for both cell number and cell size by using the Moxi Z Cell Analyzer (Orflo Technologies, Ketchum, Idaho). Cell growth was presented as the percentage of cell numbers relative to scramble control. Average cell diameter (µm) was compared in mimic-transfected versus scramble-transfected HASM cells. Data (means ± SEs) were obtained from 3 independent experiments using the same cell line. MiRNAs in airway smooth muscle (ASM) cells were sequenced by small RNA-seq, as previously described.34
RESULTS
Cohort characteristics
Of 1,041 children with asthma from the CAMP cohort, 492 (47%) had small RNA-seq data on baseline serum level available. Of these 492 CAMP participants, 448 (91%) had sufficient longitudinal FEV1 data to be classified into 1 of 4 lung growth patterns (NG [n = 137], RG [n = 120], ED [n = 102], and RGED [n = 89]) (Table I). At baseline, subjects in the RG group were more likely than participants with other patterns to be male and have a lower BMI. Subjects with either ED pattern (ED or RGED) were more likely to be slightly older and taller (8.4 and 8.5 years vs 9.1 and 9.5 years [P < .001]). Subjects with either RG pattern (RG or RGED) had worse lung function at baseline, as measured by FEV1 (88% and 85% vs 100% and 101% of predicted [P < .001]), and FVC (101% and 99% vs 108% and 109% of predicted [P < .001]), as well as more reactive airways (log PC20 [provocative concentration of methacholine causing a 20% drop in FEV1 from baseline] values of −.08 and −.23 vs .28 and .29 [P = .001]). The patterns did not differ significantly by race. The results were consistent with previous observations about these lung function patterns.10
TABLE I.
Baseline epidemiologic and clinical characteristics
| Characteristic | NG (n = 137) | ED (n = 102) | RG (n = 120) | RGED (n = 89) | P value |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 73 (53.3%) | 59 (57.8%) | 89 (74.2%) | 52 (58.4%) | .005 |
| Female | 64 (46.7%) | 43 (42.2%) | 31 (25.8%) | 37 (41.6%) | .005 |
| Age (y) | <.001 | ||||
| Mean (SD) | 8.42 (2.02) | 9.11 (2.11) | 8.51 (2.11) | 9.50 (2.10) | |
| Median [min, max] | 8.12 [5.18, 13.1] | 9.03 [5.44, 13.2] | 8.17 [5.18, 12.7] | 9.53 [5.27, 13.1] | |
| Race | .504 | ||||
| White | 103 (75.2%) | 77 (75.5%) | 99 (82.5%) | 68 (76.4%) | |
| Black | 29 (21.2%) | 22 (21.6%) | 16 (13.3%) | 15 (16.9%) | |
| Hispanic | 5 (3.6%) | 3 (2.9%) | 5 (4.2%) | 6 (6.7%) | |
| Height (cm) | <.001 | ||||
| Mean (SD) | 131 (13.0) | 135 (14.2) | 131 (13.3) | 138 (13.8) | |
| Median [min, max] | 130 [108, 170] | 136 [108, 170] | 131 [101, 167] | 137 [108, 177] | |
| BMI (kg/m2) | .019 | ||||
| Mean (SD) | 17.9 (3.15) | 18.7 (3.51) | 17.3 (2.94) | 18.2 (3.22) | |
| Median [min, max] | 17.2 [13.5, 34.3] | 17.7 [13.6, 29.7] | 16.4 [13.0, 27.4] | 17.5 [13.4, 30.7] | |
| FEV1PP | <.001 | ||||
| Mean (SD) | 100 (11.7) | 101 (11.5) | 88.1 (10.5) | 85.4 (12.5) | |
| Median [min, max] | 99.0 [64.0, 128] | 101 [65.0, 128] | 87.0 [64.0, 118] | 85.5 [54.0, 122] | |
| FVCPP | <.001 | ||||
| Mean (SD) | 108 (11.9) | 109 (11.8) | 101 (12.3) | 99.1 (12.3) | |
| Median [min, max] | 108 [71.0, 136] | 107 [86.0, 138] | 101 [69.0, 143] | 99.0 [73.0, 133] | |
| PC20 | .001 | ||||
| Mean (SD) | 0.276 (1.11) | 0.286 (1.13) | −0.0840 (1.00) | −0.234 (1.18) | |
| Median [min, max] | 0.170 [−2.08, 2.49] | 0.380 [−2.53, 2.53] | 0.0550 [−2.31, 2.22] | −0.380 [−2.94, 2.47] |
BMI, Body mass index; FEV1PP, forced expiratory volume in 1 second percent predicted; FVCPP, forced vital capacity percent predicted; PC20, provocation concentration causing a 20% fall in FEV1 value.
The subjects included in the analysis were reflective of the total CAMP population (see Table E1 in this article’s Online Repository at www.jacionline.org). Subjects were excluded only if they lacked miRNA sequencing data or a classifiable lung function pattern.
Identification of differentially expressed miRNAs
After QC, filtering, and normalization, we had a total of 259 unique miRNAs from 448 subjects for differential analysis between the 4 growth patterns. DESeq2 was used to perform normalization before differential expression (DE) analysis. Sequencing was performed in 22 batches, which may have introduced technical causes of discrepancy during preparation and handling, affecting the outcomes. Therefore, generalized principal component analysis was used to check for batch effects on mapped read counts per sample; the analysis showed that there was not a significant batch effect in normalized data (P = .931 [see Fig E1 in this article’s Online Repository at www.jacionline.org]). A total of 6 pairwise pattern conditions (NG to ED, NG to RG, NG to RGED, ED to RG, ED to RGED, and RG to RGED) were analyzed for differential miRNA expression. The complete results are listed in Table E2 (available in this article’s Online Repository at www.jacionline.org), in which the first condition is the reference group, the second condition is the comparison group, and upregulated or downregulated refers to the direction of change in the comparison group. In the ED to RG comparison, 10 miRNAs were upregulated and 10 miRNAs were downregulated; in the ED to RGED comparison, 9 miRNAs were upregulated, and 13 miRNAs were downregulated; and in the RG to RGED comparison, 2 miRNAs were upregulated and 3 miRNAs were downregulated. In the comparison of the normal group with the most severe group (ie, NG to RGED), we found only 1 (strongly) downregulated miR, namely, hsa-miR-145-5p (log2 fold change = −0.828; P < 8.01E–5; FDR = 0.021) (Fig 1). miR-145-5p was also among the upregulated miRNAs in the ED to RG condition (log2 fold change = 0.621; P = .003, FDR P = .045) and downregulated in the RG to RGED condition (log2 fold change = −0.854 [P value = .0001]; FDR P = .010). No miRNAs were differentially expressed in the comparisons of the NG to ED condition or NG to RG condition. A clustered heat map of the 20 unique differentially expressed miRs is shown in Fig 2. This indicates that miR-145-5p has expression generally orthogonal to the other differentially expressed miRs and suggests it for closer follow-up.
FIG 1.
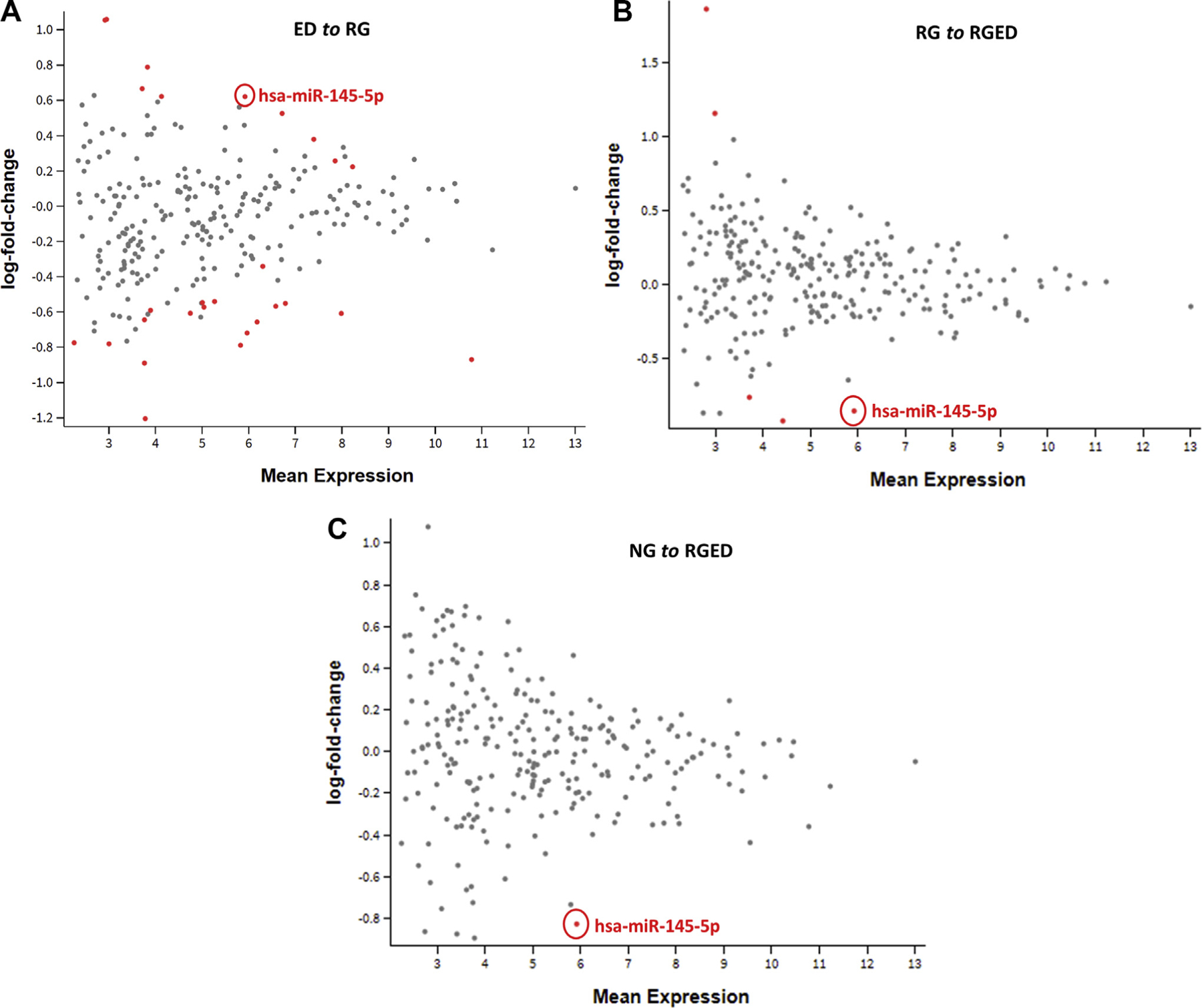
miR-145-5p is significantly differentially expressed. MD plot of ED to RG (log fold change = 0.621; P =.003, FDR P =.045) (A), RG to RGED (log fold change = −0.854; P = .0001; FDR P = .010) (B), and NG to RGED (log fold change = −0.828; P < 8.01E–05; FDR P =.021) (C).
FIG 2.
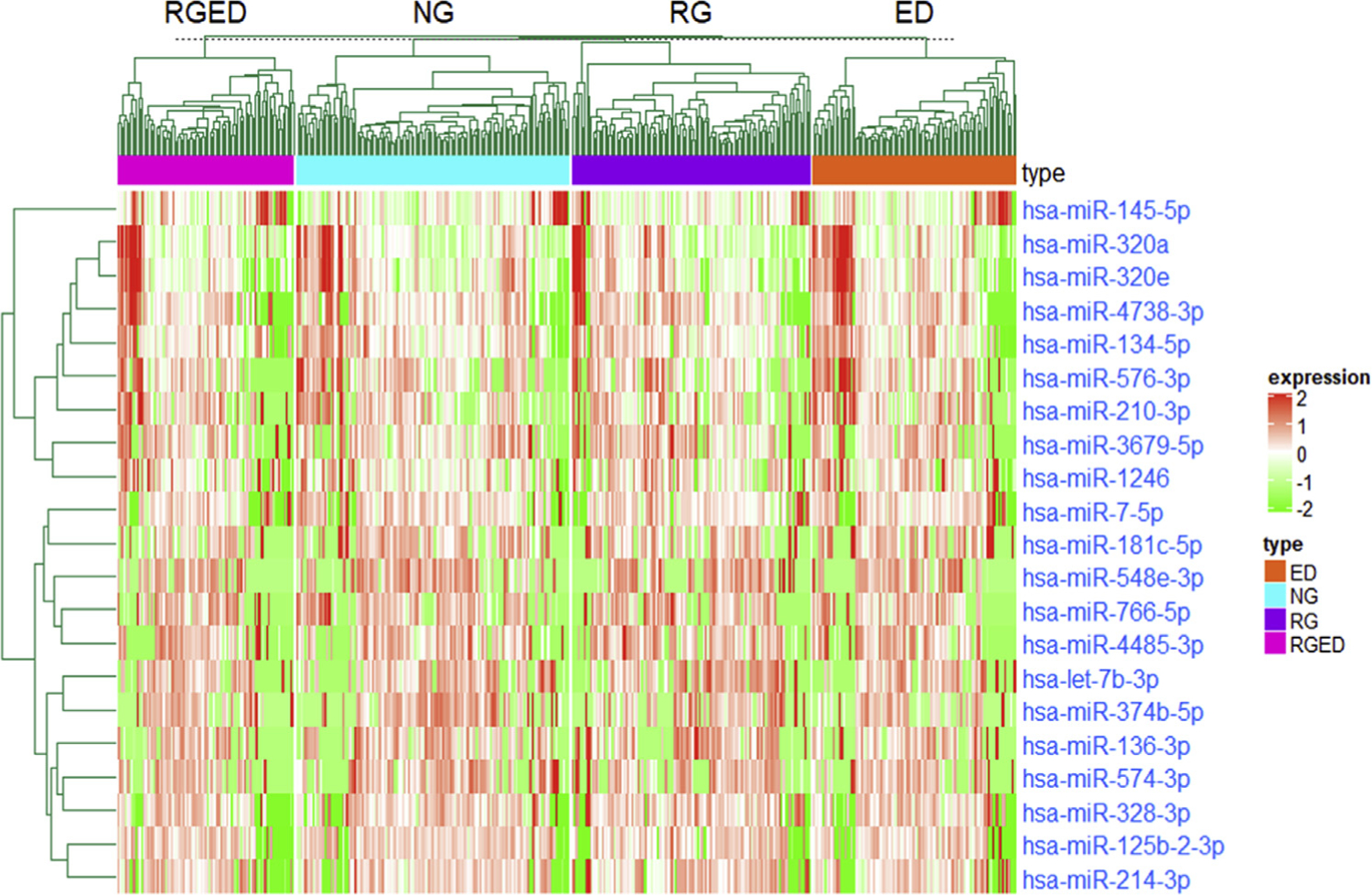
Clustered heat map of all 21 differentially expressed miRs across conditions in CAMP. Unsupervised hierarchic clustering was used to generate the heatmap, and Pearson correlation was used as the distance metric.
Association of miR-145-5p with COPD
We then prioritized miR-145-5p for deeper analysis. miR-145-5p is the mature cleaved miR of 22 nucleotides coming from the 5′ arm of the primary, hairpin, mir-145 (lowercase r). miR-145-5p and miR-145–3p were detected in the CAMP sera in similar amounts, with miR-145-5p being lower on average and with mean normalized counts of 7.52 versus 8.14 for miR-145–3p and some moderate correlation between them (r2 = 0.33).
Because miR-145-5p was associated with longitudinal patterns of lung function that have been shown to lead to COPD,10,35,36 we also checked the association of miR-145-5p with COPD at the end of follow-up (when the subjects were aged 23–30 years). There were 321 subjects (65%) with at least 1 FEV1 assessment at or beyond 23 years of age (full characterization shown in Table E3), after normalization and filtering as already described. A total of 5 miRNAs were upregulated and 25 miRNAs were downregulated between the subjects without COPD and those with COPD. We found that miRNA hsa-miR-145-5p (n = 71 with COPD and 250 without COPD; log2 fold change = −0.969; P < 5.13E–06; FDR P = .0013) was strongly underexpressed in subjects with COPD (Tables E3 and E4).
To check the ability of miR-145-5p to predict COPD at the end of follow-up, we used a linear regression model and obtained an AUC of 61% (95% CI = 56%−66%), indicating a predictive performance statistically better than random guessing. For comparison purposes, we also used all 21 differentially expressed miRs in a regression model, obtaining an AUC of 73% (95% CI = 69%−78%) (see Fig E3 in this article’s Online Repository at www.jacionline.org). However, 5-fold cross-validation indicated some overfitting and much-reduced performance of this model on a hypothetical independent validation cohort, with an AUC of 55% (95% CI = 49%−61%). In comparison, the CV of just miR-145-5p indicated performance significantly better than random (AUC = 61% [95% CI = 57%−64%]) on a hypothetical validation cohort. Accordingly, we focused much of our further analysis specifically on miR-145-5p.
Stratified analyses
CAMP was a clinical trial of 4 years of daily administration of budesonide (an inhaled corticosteroid), nedocramil, or placebo.13 Although the treatment group did not have a significant effect on the longitudinal lung function patterns,10 to assess whether the treatment group had any effect on our results we performed DE analysis by using the treatment group as a covariate, with 169 of our subjects randomized to receive budesonide, 251 randomized to receive placebo, and 28 randomized to receive nedocromil (this group was excluded from further analysis). We found that miR-145-5p was still significantly downregulated in the NG to RGED and RG to RGED pattern conditions and upregulated in the ED to RG pattern condition, as before. To assess whether miR-145-5p was more effective in particular treatments, we performed a stratified analysis by treatment group (budesonide vs placebo), finding that miR-145-5p was no longer significantly differentially expressed, although these comparisons had much-reduced sample sizes (the ED to RG comparison had 38 vs 47 samples, with the results of the other comparisons similarly reduced), and therefore reduced power. The full DE results are in Table E6, A and B (in this article’s Online Repository at www.jacionline.org).
In previous work, Howrylak et al37 identified 5 asthma subtypes within CAMP, ranging roughly from least severe (cluster 1) to most severe (cluster 5) on the basis of clinical observations. We performed a stratified analysis based on these clusters for miR-145-5p, finding that cluster 4 (moderately atopic asthma with high levels of obstruction and high exacerbation rates) retained 2 of the miR-145-5p DE results, for the NG to RGED and RG to RGED pattern conditions, while adding another for the ED to RGED pattern condition. Cluster 5 (highly atopic asthma with high levels of obstruction and high exacerbation rates) retained the result for upregulation of miR-145-5p in the ED to RG pattern condition. Other clusters did not show significant DE. The full results are in Table E6, C–G.
Putative target identification and functional enrichment analysis of differentially expressed miRNA
We retrieved a total of 80 putative gene targets for miR-145-5p from the DIANA miTG score of 0.9 or higher (see Table E5 in this article’s Online Repository at www.jacionline.org). Subsequently, these putative target genes were analyzed for the enrichment of the KEGG and GO pathways by using DAVID. The most enriched pathway cluster was characterized by the Pleckstrin homology (PH) domain (IPR001849) InterPro protein functional group (Table II), and the second most enriched cluster was characterized by the Wnt signaling pathway (KEGG hsa04310 and GO: 0016055). For comparison, we also used other providers of putative miRNA target genes. These were TargetScan, Tarbase, and miR-Walk. Fig E2 shows the overlap of these predicted targets; micro T-CDS was the most conservative and was thus likely to include the fewest false positives. PH domains were also highly enriched among TargetScan- and miRWalk-predicted targets of miR-145-5p. Cadherins were the most enriched cluster among miRWalk-predicted targets and second among Tarbase targets. Complete enrichment results are presented in Table E7, A–D (in this article’s Online Repository at www.jacionline.org).
TABLE II.
Functional enrichment of putative target genes of hsa-miR-145-5p from DAVID
| Cluster/enrichment | Functional category | ID/functional term | Gene count | Genes | % | P value | Fold | FDR |
|---|---|---|---|---|---|---|---|---|
| Annotation cluster-1 | InterPro | IPR001849 | 5 | ENSG00000116903 | 6.410 | .022 | 4.627 | 0.994 |
| Enrichment score: 1.643 | PH domain | ENSG00000106070 | ||||||
| ENSG00000115825 | ||||||||
| ENSG00000206052 | ||||||||
| ENSG00000114993 | ||||||||
| InterPro | IPR011993 | 6 | ENSG00000116903 | 7.692 | .026 | 3.524 | 0.956 | |
| PH-like domain | ENSG00000115109 | |||||||
| ENSG00000106070 | ||||||||
| ENSG00000115825 | ||||||||
| ENSG00000206052 | ||||||||
| ENSG00000114993 | ||||||||
| Annotation cluster-2 | KEGG pathway | hsa04310 Wnt signaling | 5 | ENSG00000178585 | 6.410 | .0045 | 7.121 | 0.360 |
| Enrichment score: 1.530 | pathway | ENSG00000138814 | ||||||
| ENSG00000113558 | ||||||||
| ENSG00000162337 | ||||||||
| ENSG00000163904 | ||||||||
| BP | GO: 0016055 | 4 | ENSG00000178585 | 5.128 | .040 | 5.205 | 0.996 | |
| Wnt signaling pathway | ENSG00000113558 | |||||||
| ENSG00000162337 | ||||||||
| ENSG00000163904 |
BP, Biologic process; ID, identifier.
DIANA micro T-CDS Targets of miR-145-5p were assessed for the enrichment of GWAS-identified genes for asthma, COPD, and pulmonary function by using the National Human Genome Research Institute–European Bioinformatics Institute database of GWAS results. There was significant enrichment for COPD genes (P = .018) and pulmonary function genes (P = 8.3 × 104) among the targets of miR-145-5p, whereas enrichment for asthma genes was close but nonsignificant (P = .067).
We additionally performed a pathway enrichment analysis of the target genes of all 30 differentially expressed miRNAs across all lung function patterns. Putative target genes were retrieved (764 in total; miTG score ≥ 0.99) from micro T-CDS for these 30 differentially expressed miRNAs. The top 2 clusters were characterized by transcription regulation (GO: 0006355), and by cadherins and cell adhesion (GO: 0007155), respectively (see Table E6).
In vitro validation analysis
HASM cells are an important cell type for asthma and COPD because they are in part responsible for airway remodeling, in turn leading to reduced lung function.38 Previous work by Liu et al39 showed that miR-145 had effects on HASM cells stimulated with cytokines, which could lead to airway remodeling in asthma. To test the effect of miR-145-5p on unstimulated HASM cells, we dosed cells with miR-145-5p mimics.38 We transfected mimics of the miRs in HASM cells and examined their effects on HASM growth and hypertrophy in vitro. There was no significant effect by any of the miRs on HASM hypertrophy (data not shown). However, miR-145-5p significantly increased the number of HASM cells (P < .01 [Fig 3]), indicating potential roles for miR-145-5p in HASM cell proliferation in those with asthma.
FIG 3.
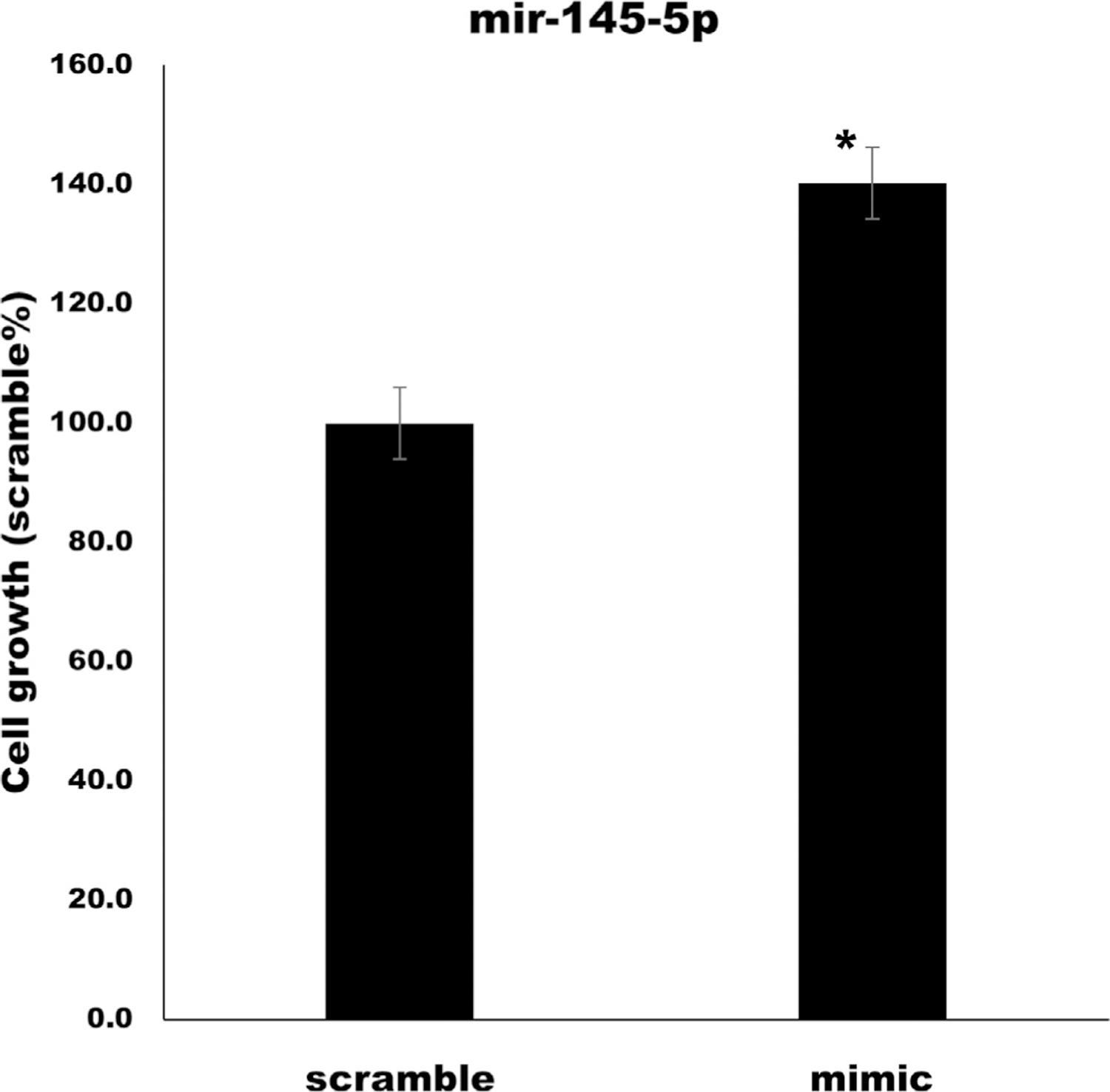
Average cell growth of HASM cells transfected with a miR-145-5p mimic versus scramble control. There were significantly more HASM cells after transfection with miR-145-5p mimic (P < .01). The SE is shown with error bars from 3 biologic replicates of each condition. SCR, Scramble control miRNA.
DISCUSSION
We have shown that circulating miR-145-5p from baseline samples in CAMP is associated with lung function trajectories over the subsequent 16 years and COPD diagnosis at that follow-up time. This represents a particularly long-term prediction. We have previously shown that miRNAs can predict long-term asthma remission40 and also predict relatively short-term asthma exacerbation rates over the following year.41
Specifically, our study demonstrated that in the comparison of the normal with the most severe group, NG to RGED, miR-145-5p was strongly downregulated, indicating less circulating miR-145-5p in the serum of participants with asthma who went on to develop very poor lung function. miR-145-5p was also upregulated in the ED to RG pattern condition and downregulated in the RG to RGED pattern condition. These comparisons overlap; together, they indicate that miR-145-5p is lower in the ED and RGED pattern condition, but higher in NG and RG pattern condition, suggesting reduced miR-145-5p as an indicator of earlier decline of lung function. The comparison of the NG and RG groups did not show significant results for miR-145-5p, but the lower expression also occurred with COPD onset before age 30 years. The effects of miR-145-5p did not seem to be related to steroid treatment but did show some increased effect in more severe asthma clusters from Howrylak et al,37 although the sample sizes in these stratified analyses were much reduced. We also observed an association of miR-145-5p with increased ASM cell proliferation. Together, these findings make a strong case for miR-145-5p as an indicator and possible cause of airway dysregulation leading to ED of lung function.
Although reduced miR-145-5p was strongly associated with declining lung function and COPD at the end of follow-up, it was not by itself a strong predictor of early COPD. It was however a robust predictor, as demonstrated by cross-validation, indicating that it should play an important part in future prognostic models of COPD and lung function in conjunction with other miRs or other genomic variants and clinical indicators.
We chose to target ASM cells because their dysregulation can lead to airway remodeling and obstructive lung disease, making them a primary tissue of interest for the transition from asthma to COPD.38 miR-145-5p is also expressed in the lung according to 2 prominent “atlases” of miRNA expression (Ludwig et al42 and McCall et al43). Indeed, according to such atlases, miR-145-5p is expressed in most, although not all, tissues. Of particular importance, in earlier cancer studies, miR-145-5p (then referred to as just miR-145) was shown to reduce cell proliferation by increasing apoptosis in a variety of tumor tissues. This occurs through a regulatory network featuring the p53 transcription factor.44–46 miR-145-5p seems to play an important role in the cell cycle and cell fate in mesenchymal stem cells as well. Preliminary work has shown that miR-145-5p and p53 work together to affect cell senescence in bone marrow,47 and to affect the transition from stem cell to ASM under certain morphologic circumstances.48 Indeed, miR-145-5p is involved in the transition of cells both to and from smooth muscle cells in other tissues. miR-145 is involved in the generation of intestinal smooth muscle cells from the mesoderm,49 and miR-145 dysregulation can also convert aortic smooth muscle cells to an aberrant form that contributes to atherosclerosis.50 These findings augment our results showing that miR-145-5p increased cell proliferation in HASM cells, as well as other results in ASM cells,39 to demonstrate the importance of miR-145-5p in a cell transition in a variety of cell types. These differences may be due to additional cell cycle dysregulation in tumor cells, or to differences in cell types, of which miRs have been shown to have different effects in different cells.51,52
Gene targets of miR-145-5p were enriched for Wnt signaling pathways. They are developmental signaling pathways that play a significant role in cell fate design, cell assimilation, and polarization. Sharma et al53 reported that Wnt signaling genes are associated with impaired lung function in childhood asthma. Another study, by Zhang et al,54 implicated Wnt/β-catenin signaling pathways in mouse models of airway remodeling in asthma. Further studies have indicated the roles of Wnt signaling in COPD pathogenesis.55,56
PH domains were enriched among miR-145-5p gene targets. PH domains are small modular domains spotted in a wide range of proteins. The domains may bind phosphatidylinositol inside biologic membranes and proteins, including the βγ-subunits of heterotrimeric G proteins and protein kinase C.57 Although some kinases have been associated with asthma, our result represents novel evidence that PH domains may play a role in ED of lung function in childhood asthma.58,59
Cadherins were enriched among gene targets of differentially expressed miRNAs across all lung growth pattern comparisons and were the most enriched cluster among miRWalk-predicted targets of miR-145-5p and second among Tarbase targets of miR-145-5p. Cadherins are transmembrane proteins that mediate cell-cell adhesion and have previously been implicated in the pathogenesis of asthma and lung disease.60,61 Cadherins have been specifically associated with the transition from airway epithelial cells to mesenchymal cells,62 resulting in airway re-modeling and reduced lung function.63,64 This provides an important link between the in silico and in vivo analyses, pointing at mechanisms of the role of miR-145-5p in the aberrant development of human ASM cells.
miR-145-5p has previously been associated with asthma, COPD, and biologic facets of both conditions. A recent study by Xiong et al demonstrated that miR-145-5p exacerbated the TH2-type immune response in mice.65 Liu et al39 investigated the effect of miR-145 on ASM function. They reported that miR-145 was highly expressed in ASM cells exposed to cytokine stimulation that mimic the airway conditions of patients with asthma. Suppression of miR-145 caused reduced ASM cell proliferation and migration in a dose-dependent manner. Consistent with our own in vitro result, Liu et al reported miR-145 overexpression in ASM cells, eventually leading to increased proliferation and migration of ASM cells in vitro.39
Childhood circulating miR-145-5p was also associated with the very early classification of COPD among CAMP participants aged 23 to 30 years at the end of follow-up. Yang et al66 found that miR-145 expression is upregulated in TGF-β1–treated lung fibroblasts and that its expression is also higher in the lungs of patients with idiopathic pulmonary fibrosis than in normal human lungs. Overexpression of miR-145 in lung fibroblasts increased α-SMA expression, enhanced contractility, and promoted the formation of focal and fibrillar adhesions. One of the most interesting results of this study is the protection of miR-145−/− mice against idiopathic pulmonary fibrosis, suggesting that miR-145-5p may be a potential target in the development of novel therapies to treat pathologic fibrotic processes that occur in the airway wall in COPD.66 Our results show that miR-145-5p level is decreased in children with asthma and ED of lung function, which may indicate a changing or complex role for miR-145-5p in determination of lung function.
Other studies have associated miR-145-5p with COPD. Wang et al demonstrated that changes in expression of miR-145-5p were associated with COPD.67 In another study, O’Leary et al reported that ASM inflammation is regulated by miR-145-5p in COPD.68 A bioinformatics-based study by Liu et al reported that miR-145 was associated with the development of COPD.69 Our results extend these findings by implicating miR-145-5p in the lung function trajectory from childhood.
Our study has several strengths. This is the first study of DE of circulating miRNA of longitudinal lung function phenotypes. Other strengths include the use of next-generation small RNA-seq to comprehensively identify miRNAs in an unbiased way. Finally, our use of an asthma cohort with at least annual spirometry allowed very accurate phenotyping of lung function trajectories.
Our study also has limitations. Primarily, we have no replication cohort for the association of miR-145-5p with longitudinal lung function patterns, owing to the difficulty in identifying detailed longitudinal FEV1 trajectories over long follow-up times. However, surveys of previous literature and functional studies provide additional evidence of the role of this miR in COPD pathogenesis. In future work, it would also be interesting to combine the short RNA-seq presented herein with (typical) long RNA-Seq to assay for mRNA transcripts, which would make it possible to obtain additional evidence of mRNA regulation by miRs. Other experiments that measure the expression of target mRNAs in vitro, as well as EdU (5-ethynyl-2′-deoxyuridine) and flow cytometry, could be performed to more accurately assess HASM cell proliferation.
Conclusion
We have shown that decreased miR-145-5p, which has previously been associated with COPD in adults, is associated with an ED of lung growth in a population of children with asthma of aged 5 to 12 years. Furthermore, a decreased baseline miR-145-5p level was also associated with COPD at the end of follow-up (at ages 23–30 years). That this miR is associated with future COPD outcomes in young children represents a significant extension of our understanding of the role of miR-145-5p in COPD. miR-145-5p was also shown to increase ASM cell proliferation. These results demonstrate that miR-145-5p and mi-RNAs in general have the potential to be powerful prognostic biomarkers for future chronic obstruction; however, additional profiling of miR-145-5p alongside longitudinal spirometry will be required.
Supplementary Material
Key messages.
Decreased miR-145-5p expression in serum is associated with early-decline longitudinal lung function from childhood to early adulthood.
miR-145-5p has previously been associated with COPD in adult populations and now seems to regulate lung function throughout the life course.
Acknowledgments
Supported by National Heart, Lung, and Blood Institute grants R01 HL139634, R01 HL127332, R01 HL129935, and P01 HL132825.
Abbreviations used
- ASM
Airway smooth muscle
- AUC
Area under the receiver-operator characteristic curve
- CAMP
Childhood Asthma Management Program
- COPD
Chronic obstructive pulmonary disease
- CV
Cross-validation
- DE
Differential expression
- ED
Early decline
- FDR
False discovery rate
- FVC
Forced vital capacity
- GO
Gene Ontology
- GWAS
Genome-wide association study
- HASM
Human airway smooth muscle cell
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- miRNA
MicroRNA
- NG
Normal growth
- PH
Pleckstrin homology
- QC
Quality control
- RG
Reduced growth
- RGED
Reduced growth with an early decline
- RNA-seq
RNA sequencing
Footnotes
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 2018;9:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hobbs BD, Tantisira KG. MicroRNAs in COPD: small molecules with big potential. Eur Respir J 2019;53. [DOI] [PubMed] [Google Scholar]
- 3.Brown D, Rahman M, Nana-Sinkam SP. MicroRNAs in respiratory disease. a clinician’s overview. Ann Am Thorac Soc 2014;11:1277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahamtan A, Teymoori-Rad M, Nakstad B, Salimi V. Anti-inflammatory micro-RNAs and their potential for inflammatory diseases treatment. Front Immunol 2018;9:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vijayan KK. Chronic obstructive pulmonary disease. Ind J Med Res 2013;137: 251–69. [PMC free article] [PubMed] [Google Scholar]
- 6.Dougherty RH, Fahy JV. Acute exacerbations of asthma: epidemiology, biology, and the exacerbation-prone phenotype. Clin Exp Allergy 2009;39: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerkhof M, Boezen HM, Granell R, Wijga AH, Brunekreef B, Smit HA, et al. Transient early wheeze and lung function in early childhood associated with chronic obstructive pulmonary disease genes. J Allergy Clin Immunol 2014;133:68–76.e4. [DOI] [PubMed] [Google Scholar]
- 8.McGeachie MJ, Clemmer GL, Croteau-Chonka DC, Castaldi PJ, Cho MH, Sordillo JE, et al. Whole-genome prediction and heritability of childhood asthma phenotypes. Immun Inflamm Dis 2016;4:487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGeachie MJ, Yates KP, Zhou X, Guo F, Sternberg AL, Van Natta ML, et al. Genetics and genomics of longitudinal lung function patterns in individuals with asthma. Am J Respir Crit Care Med 2016;194:1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGeachie MJ, Yates KP, Zhou X, Guo F, Sternberg AL, Van Natta ML, et al. Patterns of growth and decline in lung function in persistent childhood asthma. N Engl J Med 2016;374:1842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu K, Gamazon ER, Im HK, Geeleher P, White SR, Solway J, et al. Genome-wide interrogation of longitudinal FEV1in children with asthma. Am J Respir Crit Care Med 2014;190:619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res 2011;717:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 14.Cleveland WS. LOWESS: a program for smoothing scatterplots by robust locally weighted regression. Am Stat 1981;35:54. [Google Scholar]
- 15.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med 1999;159:179–87. [DOI] [PubMed] [Google Scholar]
- 16.Matias-Garcia PR, Wilson R, Mussack V, Reischl E, Waldenberger M, Gieger C, et al. Impact of long-term storage and freeze-thawing on 8 circulating microRNAs in plasma samples. PLoS One 2020;15:e0227648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balzano F, Deiana M, Dei Giudici S, Oggiano A, Baralla A, Pasella S, et al. miRNA stability in frozen plasma samples. Molecules 2015;20:19030–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozowsky J, Kitchen RR, Park JJ, Galeev TR, Diao J, Warrell J, et al. exceRpt: a comprehensive analytic platform for extracellular RNA profiling. Cell Syst 2019;8: 352–7.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese SE, Archer KJ, Therneau TM, Atkinson EJ, Vachon CM, de Andrade M, et al. A new statistic for identifying batch effects in high-throughput genomic data that uses guided principal component analysis. Bioinformatics 2013;29:2877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubitzyk W, Wolkenhauer O, Cho K-H, editors. Encyclopedia of system biology. Springer; 2013. pp. 2367. [Google Scholar]
- 22.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:847–9. [DOI] [PubMed] [Google Scholar]
- 23.Provost F, Fawcett T. Robust classification for imprecise environments. Mach Learn 2001;44:203–31. [Google Scholar]
- 24.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–45. [PubMed] [Google Scholar]
- 25.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 2013;41:W169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein-coding sequences. Bioinformatics 2012;28:771–6. [DOI] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2008;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim, and improvements in enrichment analysis tools. Nucleic Acids Res 2018;47:D419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2016;45: D353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 2017; 45:D190–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018;46:D239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu R, Pan W, Fedulov AV, Jester W, Jones MR, Weiss ST, et al. MicroRNA-10a controls airway smooth muscle cell proliferation via direct targeting of the PI3 kinase pathway. FASEB J 2014;28:2347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bui DS, Lodge CJ, Burgess JA, Lowe AJ, Perret J, Bui MQ, et al. Childhood predictors of lung function trajectories and future COPD risk: a prospective cohort study from the first to the sixth decade of life. Lancet Respir Med 2018;6:535–44. [DOI] [PubMed] [Google Scholar]
- 36.McGeachie MJ. Childhood asthma is a risk factor for the development of chronic obstructive pulmonary disease. Curr Opin Allergy Clin Immunol 2017;17:104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howrylak JA, Fuhlbrigge AL, Strunk RC, Zeiger RS, Weiss ST, Raby BA, et al. Classification of childhood asthma phenotypes and long-term clinical responses to inhaled anti-inflammatory medications. J Allergy Clin Immunol 2014;133: 1289–300.e1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazaar AL, Panettieri RA Jr. Airway smooth muscle: a modulator of airway remodeling in asthma. J Allergy Clin Immunol 2005;116:488–95, quiz 96. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Sun X, Wu Y, Fang P, Shi H, Xu J, et al. Effects of miRNA-145 on airway smooth muscle cells function. Mol Cell Biochem 2015;409:135–43. [DOI] [PubMed] [Google Scholar]
- 40.McGeachie MJ, Davis JS, Kho AT, Dahlin A, Sordillo JE, Sun M, et al. Asthma remission: predicting future airways responsiveness using an miRNA network. J Allergy Clin Immunol 2017;140:598–600.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kho AT, McGeachie MJ, Moore KG, Sylvia JM, Weiss ST, Tantisira KG. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir Res 2018;19:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res 2016;44: 3865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCall MN, Kim MS, Adil M, Patil AH, Lu Y, Mitchell CJ, et al. Toward the human cellular microRNAome. Genome Res 2017;27:1769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol 2011;3:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A 2009;106:3207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Sun Z, Yan S, Gao F. Effect of miR145 on gastric cancer cells. Mol Med Rep 2019;19:3403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia C, Jiang T, Wang Y, Chen X, Hu YH, Gao Y. The role of p53-induced miR-145a in senescence and osteogenesis of mesenchymal stem cells. Research Square preprint 2020.
- 48.Yeh YT, Wei J, Thorossian S, Nguyen K, Hoffman C, Del Alamo JC, et al. MiR-145 mediates cell morphology-regulated mesenchymal stem cell differentiation to smooth muscle cells. Biomaterials 2019;204:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gays D, Hess C, Camporeale A, Ala U, Provero P, Mosimann C, et al. An exclusive cellular and molecular network governs intestinal smooth muscle cell differentiation in vertebrates. Development 2017;144:464–78. [DOI] [PubMed] [Google Scholar]
- 50.Vengrenyuk Y, Nishi H, Long X, Ouimet M, Savji N, Martinez FO, et al. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler Thromb Vasc Biol 2015;35:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J Mol Cell Cardiol 2012;52:245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsin JP, Lu Y, Loeb GB, Leslie CS, Rudensky AY. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat Immunol 2018;19:1137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma S, Tantisira K, Carey V, Murphy AJ, Lasky-Su J, Celedon JC, et al. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am J Respir Crit Care Med 2010;181:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang W, Zhu T, Jia X, Weng C, Li C, Zhao W. Wnt7b/beta-catenin signal pathway associated with airway remodeling of asthma rats. J Allergy Clin Immunol 2017; 139:AB3. [Google Scholar]
- 55.Baarsma HA, Skronska-Wasek W, Mutze K, Ciolek F, Wagner DE, John-Schuster G, et al. Noncanonical WNT-5A signaling impairs endogenous lung repair in COPD. J Exp Med 2017;214:143–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skronska-Wasek W, Mutze K, Baarsma HA, Bracke KR, Alsafadi HN, Lehmann M, et al. Reduced frizzled receptor 4 expression prevents WNT/beta-catenin-driven alveolar lung repair in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2017;196:172–85. [DOI] [PubMed] [Google Scholar]
- 57.Wang DS, Shaw R, Winkelmann JC, Shaw G. Binding of PH domains of beta-adrenergic receptor kinase and beta-spectrin to WD40/beta-transducin repeat containing regions of the beta-subunit of trimeric G-proteins. Biochem Biophys Res Commun 1994;203:29–35. [DOI] [PubMed] [Google Scholar]
- 58.Yao L, Suzuki H, Ozawa K, Deng J, Lehel C, Fukamachi H, et al. Interactions between protein kinase C and pleckstrin homology domains. Inhibition by phosphatidylinositol 4,5-bisphosphate and phorbol 12-myristate 13-acetate. J Biol Chem 1997;272:13033–9. [DOI] [PubMed] [Google Scholar]
- 59.Barnes PJ. Kinases as novel therapeutic targets in asthma and chronic obstructive pulmonary disease. Pharmacol Rev 2016;68:788–815. [DOI] [PubMed] [Google Scholar]
- 60.Post S, Heijink IH, Hesse L, Koo HK, Shaheen F, Fouadi M, et al. Characterization of a lung epithelium specific E-cadherin knock-out model: implications for obstructive lung pathology. Sci Rep 2018;8:13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maître J-L, Heisenberg C-P. Three functions of cadherins in cell adhesion. Curr Biol 2013;23:R626–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Z, Ji N, Ma Q, Zhu R, Chen Z, Wang Z, et al. Epithelial-mesenchymal transition in asthma airway remodeling is regulated by the IL-33/CD146 axis. Front Immunol 2020;11:1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Muhsen S, Johnson JR, Hamid Q. Remodeling in asthma. J Allergy Clin Immunol 2011;128:451–62, quiz 63–4. [DOI] [PubMed] [Google Scholar]
- 64.Koppelman GH, Meyers DA, Howard TD, Zheng SL, Hawkins GA, Ampleford EJ, et al. Identification of PCDH1as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med 2009;180:929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiong T, Du Y, Fu Z, Geng G. MicroRNA-145-5p promotes asthma pathogenesis by inhibiting kinesin family member 3A expression in mouse airway epithelial cells. J Int Med Res 2019;47:3307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, et al. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J 2013;27:2382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang M, Huang Y, Liang Za, Liu D, Lu Y, Dai Y, et al. Plasma miRNAs might be promising biomarkers of chronic obstructive pulmonary disease. Clin Respir J 2014;10:104–11. [DOI] [PubMed] [Google Scholar]
- 68.O’Leary L, Sevinc K, Papazoglou IM, Tildy B, Detillieux K, Halayko AJ, et al. Airway smooth muscle inflammation is regulated by microRNA-145 in COPD. FEBS Lett 2016;590:1324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Qu J, Xue W, He L, Wang J, Xi X, et al. Bioinformatics-based identification of potential microRNA biomarkers in frequent and non-frequent exacerbators of COPD. Int J Chron Obstruct Pulmon Dis 2018;13:1217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


