Abstract
Live Attenuated Influenza Virus (LAIV) is administered to and replicates in the sinonasal epithelium. Candidate LAIV vaccine strains are selected based on their ability to replicate to a high titer in embryonated hen’s eggs, a process that can lead to mutations which alter the receptor binding and antigenic structure of the hemagglutinin (HA) protein. In the 2012–2013 northern hemisphere vaccine, the H3N2 HA vaccine strain contained three amino acid changes - H156Q, G186V and S219Y – which altered HA antigenic structure and thus presumably decreased vaccine efficacy. To determine if these mutations also altered LAIV replication, reabcombinant viruses were created that encoded the wild-type (WT) parental HA of A/Victoria/361/2011 (WT HA LAIV), the egg adapted HA (EA HA LAIV) from the A/Victoria/361/2011 vaccine strain and an HA protein with additional amino acid changes to promote α2,3 sialic acid binding (2,3 EA HA LAIV). The WT HA LAIV bound α2,6 sialic compared to the EA HA LAIV and 2,3 EA HA LAIV which both demonstrated an increased preference for α2,3 sialic acid. On MDCKs, the WT HA and EA HA LAIVs showed similar replication at 32°C but at 37°C the EA HA LAIV replicated to lower infectious virus titers. The 2,3 EA HA LAIV replicated poorly at both temperatures. This replication phenotype was similar on human nasal epithelial cell (hNEC) cultures, however the WT HA LAIV induced the highest amount of IFN-λ and infected more nasal epithelial cells compared to the other viruses. Together, these data indicate that egg adaption mutations in the HA protein that confer preferential α2,3 sialic acid binding may adversely affect LAIV replication and contribute to reduced vaccine efficacy.
Keywords: influenza, nasal epithelial cells, LAIV, vaccines, egg adaption
Introduction
Influenza A virus (IAV) is a member of the Orthomyxoviridae family of viruses. Each year IAV infects between 10 and 20% of the world population, causes thousands of deaths and results in millions of dollars in economic losses (1). There are options available for annual influenza vaccination, one of which is the live, attenuated influenza virus (LAIV) vaccine. The LAIV is a cold adapted and attenuated IAV. The LAIV strain used in the United States was generated by serially passaging a seasonal H2N2 virus, A/Ann Arbor/6/1960, in embryonated hen’s eggs at progressively cooler temperatures (2–5). The resulting virus was capable of growth at lower temperature (25°C), and was attenuated at 37°C and 39°C in animal models (6). There is a similarly attenuated LAIV donor strain, A/Leningrad/134/17/57 H2N2, used in other parts of the world (7).
LAIV replicates in the cooler environment of the upper airways but has restricted growth in the warmer lower airways. The temperature attenuation phenotype has been mapped to mutations in the polymerase complex (PA, PB2, PB1 and NP) as well as the matrix protein M2 (2, 8). The mutations result in inefficient viral RNA synthesis, defective particle formation, deficient replication, and a reduced incorporation of the viral matrix M1 protein into progeny virions (6, 9, 10). LAIV produces the same amount of viral particles at 37°C compared to a matched wild type (WT) virus, but the ratio of infective particles to total particles is significant lower (11).
The LAIV vaccine is a recombinant 6:2 reassorted virus created by selecting the hemagglutinin (HA) and neuraminidase (NA) gene segments from circulating seasonal influenza viruses and rescuing virus with the remaining gene segments derived from a master donor LAIV strain. This results in a virus that displays the surface glycoproteins from circulating IAV strains, replicates in the upper airway, stimulates an innate, humoral and cellular immune response but fails to replicate efficiently in the lower airway (12, 13).
Both the inactivated influenza vaccine (IIV) and LAIV are grown in embryonated hen’s eggs, partially purified, then administered in their respective vaccine formulation (14, 15). During egg propagation and selection of a candidate vaccine virus (CVV), the virus can accumulate amino acid mutations in the HA protein (16, 17). Most of these changes are in the receptor binding site located in the head of HA and often overlap with antigenic regions. Increased replication leads to higher per egg yields of vaccine (16, 18). Egg propagated viruses can undergo antigenic changes. One notable instance was in the 2012–13 season where the H3N2 vaccine component (A/Victoria/361/2011) was antigenically mismatched from circulating viruses that season (19). Other studies have suggested the H3 IIV component was also poorly immunogenic but induced an antibody response that did recognize circulating H3 strains (20). In either case, the effects of mutations in HA may have greater consequence for LAIV, since this virus must replicate and mimic a natural influenza infection – something not required of IIV. While LAIV’s are historically tested for replication competence in immortalized cells, very little research has been conducted on the ability of H3N2 LAIVs to replicate in human nasal epithelial cells and how egg adaption of the LAIV HA receptor-binding impacts LAIV fitness and efficacy (21–23).
We found that egg adaptation of the H3N2 LAIV vaccine strain for the 2012/13 season results in sialic acid receptor binding changes, decreased replication efficiency on human nasal epithelial cell (hNECs) cultures, decreased innate immune induction and a decreased efficiency of infecting hNEC cultures. Switching receptor preference by producing the LAIV vaccine in eggs could potentially have a dramatic effect on the replication and vaccine efficacy of the LAIV vaccine platform.
Materials and Methods
Cell Lines and Primary Cells
Madin-Darby Canine Kidney Cells (MDCK) and human embryonic kidney cells 293T (HEK293T) were maintained in complete medium (CM) consisting of Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 100U/ml penicillin/streptomycin (Life Technologies) and 2mM Glutamax (Gibco). Human nasal epithelial cells (hNEC) were isolated from non-diseased donor tissue following endoscopic sinus surgery. Cells were grown in Trans-well 24 well plates, differentiated and maintained at the air liquid interface (ALI) as previously described (6, 8, 24). hNEC differentiation medium and maintenance medium was prepared as previously described (6, 13, 25). The hNEC cultures were used for low multiplicity of infection (MOI) growth curves only when fully differentiated. All cells were maintained at 37°C in a humidified incubator supplemented with 5% CO2. hNEC cultures were acclimated to 32°C for 48 hours before infection.
Plasmids
The plasmid pHH21 was used to generate full length influenza hemagglutinin (HA) or neuraminidase (NA) gene segment encoding plasmids for recombinant virus production. LAIV internal segments from the cold adapted A/Ann Arbor/6/1960 were used as previously described (6, 8, 11). Briefly, viral RNA was isolated from A/Victoria/361/2011 virus stock solutions with a Qiagen mini-vRNA isolation kit. Gene specific primers with cloning sites for the A/Victoria/361/2011 (H3N2) neuraminidase or hemagglutinin (GISAID Accession no.: EPI_ISL_134450) were used to create cDNA via a one-step RT-PCR reaction (SuperScript III-Platinum Taq mix, ThermoFisher Scientific). The RT-PCR products were cut with appropriate restriction enzymes, column purified (QIAquick PCR Purification kit) and ligated with restriction enzyme cut-pHH21 using T4-ligase (New England Biolabs, NEB). To create the EA and 2,3 EA HA plasmids, site directed mutagenesis was performed on the WT plasmid (Agilent). Three separate mutations were performed on the WT HA plasmid (H156Q, G186V and S219Y) to create the EA HA plasmid, per the available IVR-165 sequence (26). To create the 2,3 EA HA plasmid, two further amino acid mutations were introduced into the EA HA plasmid via site directed mutagenesis (I226Q and S228G). All plasmids were maxi prepped and Sanger sequence verified before use in recombinant virus production.
Recombinant Virus Production
Recombinant H3N2 LAIV viruses were generated using the 12 plasmid reverse genetics system as previously described (27, 28). For recombinant virus seed stocks, 250μl of media from one picked plaque was added to confluent MDCK cells plated in 6 well plates and infected for 1hr as previously described (24, 29). The HA protein sequence was determined from working stocks to confirm the presence of the introduced mutations.
Plaque Assay and TCID50
Both the plaque assay and TCID50 assays for determining infectious virus production were performed as described previously (27, 28).
Virus seed and working stocks
Virus Particle Purification
Approximately 150ml of supernatant from virus-infected cells was clarified by centrifugation at 300g for 10 minutes, then overlaid onto a 25% sucrose-NTE (100nM NaCl (ThermoFisher Scientific), 10mM Tris (Promega) and 1mM EDTA (Sigma)) buffer. The samples were centrifuged at 27,000 RPM in a SW-28 rotor in a Beckman Coulter Optima L90-K UltraCentrifuge for 2 hours. The supernatant was removed, the virus pellet was re-suspended in PBS and further concentrated by ultracentrifugation in an SW-28ti rotor at 23,000 RPM for 1hr. The pellet was resuspended in PBS. Partially purified virus particles were then labeled with Alexa Fluor 488 Succinimidyl Ester per the manufacturer’s instructions (Thermofisher Scientific).
Consortium for Functional Glycomics Glycan Array
To assess HA receptor specificity, partially purified, whole virus particles labeled with fluorescent dye were allowed to bind to Consortium for Functional Glycomics Array version 5. synthetic glycan chip as previously described (30–33). Labeled virus was allowed to bind to an array chip for 1hr at room temperature, then excess was aspirated. Slides were washed three times before fluorescence analysis. The array chip was scanned with GenePix 4300A Microarray scanner, data was analyzed with GenePix Pro Microarray Analysis Software and processed via Excel spreadsheets as previously described (30–33). Only sialic acid containing ligands were considered for analysis but a complete list of glycans is available on the CFG website (34)
Low-MOI Infections
Low-MOI growth curves were performed at an MOI of .001 infectious units/cell in MDCK cells and .015 in hNEC cultures. MDCK cell infections were performed as described above. After the infection, the inoculum was removed and the MDCK cells were washed three times with PBS+. After washing, IM was added and the cells were placed at 32°C. At the indicated times post inoculation, IM was removed from the MDCK cells and frozen at −80°C. Fresh IM was then added. In low-MOI hNEC growth curves, hNECs were acclimated to 32°C or 37°C for 48hrs before infection. The apical surface was washed three times with PBS and the basolateral media was changed at time of infection. hNEC cultures were inoculated at an MOI of 0.015. hNEC cultures were then placed in a 32°C incubator for 2 hours. After incubation, the apical surface of the hNEC culture was washed three times with PBS. At the indicated times, 100ul of IM without N-acetyl trypsin was added to the apical surface of the hNECs for 5 minutes at 32°C, the IM was harvested and stored at −80°C. Basolateral media was changed every 24hrs post infection for the duration of the experiment.
Basolateral IFN-λ Analysis
Secreted IFN-λ was quantified from basolateral samples taken during low MOI hNEC infections taken 24 hours post infection. The DIY Human IFN-λ I/II/III (IL-29/28A/28B) ELISA (PBL Assay Science) was used according to manufacturers’ instructions. Samples were diluted 1:4 to stay within the detection range of the assay. Values of IFN-λ were adjusted by subtracting mock infected and plotted as picograms/ml.
Flow Cytometry
hNECs were infected for 14 hours at an MOI of 3 at 32°C. After infection, apical and basolateral surfaces were washed with PBS twice. Cells were then detached with 1X TrypLE (Life Technologies) for 15 minutes at 37°C and cells were pelleted by centrifugation. Cells were washed again with PBS and fixed with 2% paraformaldehyde (Affymetrix) at room temperature for 15 minutes. Cells were permeabilized with PBS containing 0.2% Tween-20 for 15 minutes at room temperature. The cells were then incubated with blocking buffer (PBS with 1% solution of serum from the same species as the secondary antibody and 0.3% BSA). After blocking, cells were incubated with indicated primary antibodies, washed twice, incubated with secondary antibodies. Each washing step was done with FACS buffer (PBS with 0.3% BSA). Cells were analyzed on a BD-LSR II and data analyzed with FlowJo V10.5.3 software.
Antibodies
Antibodies against human beta tubulin IV (Novus Bio, clone OTI3F1) conjugated to Alexa Fluor 488 were used at a concentration of 0.5μg/ml to identify ciliated cells in hNEC cultures. Goat anti-Aichi H3N2 serum (BEI resources) diluted 1:500 was used to identify infected cells. Donkey anti-goat IgG Alexa Fluor 647 (ThermoFisher Scientific), diluted to 0.5μg/ml was used to detect unconjugated anti-serum.
Results
Amino acid mutations that occur in human influenza viruses grown in embryonated hen’s eggs typically alter the sialic acid binding preference (16, 18, 35–38). To study the effect that egg adaptation has on receptor binding specificity, virus replication and innate immune induction, a panel of three recombinant LAIVs were produced. All three HA proteins are based on the 2012/2013 northern hemisphere H3N2 vaccine component, A/Victoria/361/2011. The wild-type (WT) HA was cloned from the WT A/Victoria/361/2011 HA sequence. The three amino acid modifications observed in the egg-adapted (EA) vaccine strain (26) H156Q, G186V and S219Y, were then introduced. These amino acid mutations have been previously shown to shift sialic acid binding from the human α2,6 receptor to the avian α2,3 receptor (17). Two additional amino acid changes were introduced into the EA HA gene to make the 2,3 EA HA which is predicted to bind preferentially to α2,3 sialic acid moieties. These amino acid changes, I226Q and S228G, have been shown to change sialic acid binding preference from α2,6 to α2,3 sialic acid in other H3 proteins (39). The 3D structure with highlighted amino acid changes near the sialic acid binding pocket residues of each protein is shown for the WT (Figure 1A), EA (Figure 1B) and 2,3 EA (Figure 1C) HA proteins.
Figure 1. 3D modeling of A/Victoria/361/2011 HA proteins.
(A) Model of WT A/Victoria/361/2011 HA monomer protein. Sialic acid binding pocket residues highlighted (60–64) 190 Helix in pink, 130 loop in blue and 220 loop in green. (B) Model of EA A/Victoria/361/2011 HA. Egg adaptation of A/Victoria/361/2011 during vaccine manufacturing resulted in three amino acid changes, highlighted on the structure (H156Q, G186V and S219Y). (C) Model of 2,3 EA HA. Two additional amino acid changes were added to the EA HA to shift sialic acid binding preference to α2,3 sialic acid (I226Q and S228G). Images were prepared with UCSF Chimera software with the Protein DataBase structure ID 4WE9 (A/Victoria/361/2011).
The HA proteins were successfully rescued in the LAIV genetic background and characterized for their sialic acid binding using a synthetic glycan array available from the Consortium for Functional Glycomics (v5.3 array) (34). This array contains 156 glycans that contain the human α2,6 sialic acid receptor ligand, the avian α2,3 sialic acid receptor ligand, a mixture of each, or the non-traditional α2,8 sialic acid linkage (34). The WT HA LAIV, having an identical HA sequence to the circulating human clinical isolate, bound to a number of α2,6 and α2,3 sialic acid ligands (Figure 2A, Table 1). Out of the top 20 highest binding glycans, 12 of them contained the α2,6 sialic acid ligand (Table 1). The EA HA LAIV, also bound to a mixture of α2,6 and α2,3 sialic acid containing ligands. However, out of the top 20 highest binding glycans, only 8 out of 20 contained the human α2,6 sialic acid ligand (Figure 2B, Table 2). For the 2,3 EA, 7 out of the top 20 contained the α2,6 sialic acid receptor ligand (Figure 2C, Table 3). The glycan array analysis clearly demonstrates that the amino acid differences between these HA proteins alters glycan binding preference.
Figure 2: Glycan array analysis of recombinant H3N2 LAIV’s with different HA proteins.
WT HA (A), EA HA (B) and 2,3 EA HA (C) A/Victoria/361/2011 LAIVs were subjected to glycan array analysis. The Y axis indicates raw average relative fluorescence units (RFU) while the X axis indicates the specific glycan. See Tables 1–3 for information regarding chemical structure and glycan ID. α2,3 Sialic acid containing ligands in blue box (glycans 2–68) and α2,6 sialic acid containing glycans in off white box (glycans 75–118).
Table 1:
WT HA A/Victoria/361/2011 CFG synthetic glycan array binding data
| Sample ID | Average RFU | α2,3 or α2,6 SA |
|---|---|---|
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 44628 | α2,6 |
| Neu5Aca2-6GalNAcb1-4GlcNAcb-Sp0 | 41593 | α2,6 |
| Neu5Aca2-6Galb1-4(6S)GlcNAcb-Sp8 | 36903 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-2Mana1-6(Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-2Mana1-3)Manb1-4GlcNAcb1-4GlcNAcb-Sp12 | 35455 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 34159 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 32877 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb-Sp8 | 31044 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb-Sp0 | 30317 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-6(Galb1-3)GalNAca-Sp14 | 20914 | α2,6 |
| Neu5Aca2-6GalNAcb1-4(6S)GlcNAcb-Sp8 | 15903 | α2,6 |
| Neu5Aca2-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 9754 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb1-2Mana1-6(GlcNAcb1-4)(Neu5Aca2-3Galb1-4GlcNAcb1-2Mana1-3)Manb1-4GlcNAcb1-4GlcNAcb-Sp21 | 8028 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 7644 | α2,3 |
| Galb1-4GlcNAcb1-6(Neu5Aca2-6Galb1-3GlcNAcb1-3)Galb1-4Glc-Sp21 | 4353 | α2,6 |
| Neu5Aca2-3Galb1-4GlcNAcb1-3Galb-Sp8 | 4184 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb1-2Mana1-6(Neu5Aca2-3Galb1-4GlcNAcb1-2Mana1-3)Manb1-4GlcNAcb1-4GlcNAcb-Sp12 | 4119 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb-Sp0 | 3734 | α2,3 |
| Neu5Aca2-6(Neu5Aca2-3Galb1-3)GalNAca-Sp8 | 3214 | α2,6 |
| Neu5Aca2-3Galb1-4GlcNAcb1-3Galb1-3GlcNAcb-Sp0 | 3186 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb1-6(Galb1-3)GalNAca-Sp14 | 3153 | α2,3 |
A/Victoria/361/2011 LAIV with WT HA used in CFG synthetic glycan array top 20 values. Sialic acid (SA) orientation indicated (α2,3 vs α2,6). RFU average was calculated by taking the 4 highest RFU values (out of 6 technical replicates). Out of the top 20, 12 glycans contain α2,6 SA linkages and 8 contains α2,3 SA.
Table 2:
EA HA A/Victoria/361/2011 CFG synthetic glycan array binding data
| Sample ID | Average RFU | α2,3 or α2,6 SA |
|---|---|---|
| Neu5Aca2-6GalNAcb1-4GlcNAcb-Sp0 | 43634 | α2,6 |
| Neu5Aca2-3GalNAcb1-4GlcNAcb-Sp0 | 25436 | α2,3 |
| Neu5Aca2-6Galb1-4(6S)GlcNAcb-Sp8 | 19376 | α2,6 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb-Sp8 | 17094 | α2,3 |
| Neu5Aca2-6Galb1-4GlcNAcb-Sp8 | 14351 | α2,6 |
| Neu5Aca2-6(Galb1-3)GlcNAcb1-4Galb1-4Glcb-Sp10 | 13421 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb-Sp0 | 12390 | α2,6 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 12333 | α2,3 |
| Neu5Aca2-6GalNAca-Sp8 | 10025 | α2,6 |
| Neu5Aca2-6(Neu5Aca2-3Galb1-3)GalNAca-Sp8 | 10025 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 9878 | α2,6 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb1-2Mana-Sp0 | 9869 | α2,3 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 9863 | α2,6 |
| Neu5Aca2-6GalNAcb1-4(6S)GlcNAcb-Sp8 | 9269 | α2,3 |
| Neu5Aca2-3Galb1-4(Neu5Aca2-3Galb1-3)GlcNAcb-Sp8 | 8602 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb-Sp8 | 6823 | α2,3 |
| Neu5Aca2-3Galb-Sp8 | 4391 | α2,3 |
| Neu5Aca2-3Galb1-4GlcNAcb1-6(Neu5Aca2-3Galb1-4GlcNAcb1-2)Mana1-6(GlcNAcb1-4)(Neu5Aca2-3Galb1-4GlcNAcb1-4(Neu5Aca2-3Galb1-4GlcNAcb1-2)Mana1-3)Manb1-4GlcNAcb1-4GlcNAcb-Sp21 | 4121 | α2,3 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb-Sp8 | 4032 | α2,3 |
| Neu5Gca2-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 3833 | α2,3 |
A/Victoria/361/2011 LAIV with EA HA used in CFG synthetic glycan array top 20 values. Sialic acid (SA) orientation indicated (α2,3 vs α2,6). RFU average was calculated by taking the 4 highest RFU values (out of 6 technical replicates). Out of the top 20, 8 glycans contain α2,6 SA linkages and 12 contains α2,3 SA.
Table 3:
2,3 EA HA A/Victoria/361/2011 CFG synthetic glycan array binding data
| Structure ID | Average RFU | α2,3 or α2,6 SA |
|---|---|---|
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 21866 | α2,3 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb-Sp8 | 19281 | α2,3 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb-Sp8 | 15227 | α2,3 |
| Neu5Aca2-3Galb1-3(6S)GalNAca-Sp8 | 11236 | α2,3 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 10638 | α2,6 |
| Neu5Aca2-6GalNAcb1-4GlcNAcb-Sp0 | 10557 | α2,6 |
| Neu5Aca2-6Galb1-4(6S)GlcNAcb-Sp8 | 9451 | α2,6 |
| Neu5Aca2-6(Neu5Aca2-3Galb1-3)GalNAca-Sp8 | 8016 | α2,6 |
| Neu5Aca2-6Galb1-4GlcNAcb1-3Galb1-4GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 7312 | α2,6 |
| Neu5Aca2-3Galb1-4GlcNAcb1-6(Neu5Aca2-3Galb1-3)GalNAca-Sp14 | 7025 | α2,3 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 6599 | α2,3 |
| Neu5Gca2-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 6277 | α2,3 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb1-2Mana-Sp0 | 6234 | α2,6 |
| Neu5Aca2-6(Neu5Aca2-3Galb1-3)GalNAca-Sp14 | 6220 | α2,6 |
| Neu5Aca2-3Galb1-3GlcNAcb1-3GalNAca-Sp14 | 5952 | α2,3 |
| Neu5Aca2-3Galb1-3GlcNAcb1-3Galb1-4GlcNAcb-Sp0 | 5725 | α2,3 |
| Neu5Aca2-3Galb1-3(6S)GlcNAc-Sp8 | 5359 | α2,3 |
| Neu5Aca2-3Galb1-3GalNAca-Sp8 | 5332 | α2,3 |
| Neu5Aca2-3Galb1-4(Fuca1-3)GlcNAcb1-3Galb1-4GlcNAcb-Sp8 | 4920 | α2,3 |
| Neu5Aca2-3Galb1-3(Fuca1-4)GlcNAcb1-3Galb1-4(Fuca1-3)GlcNAcb-Sp0 | 4836 | α2,3 |
A/Victoria/361/2011 LAIV with 2,3 EA HA used in CFG synthetic glycan array top 20 values. Sialic acid (SA) orientation indicated (α2,3 vs α2,6). RFU average was calculated by taking the 4 highest RFU values (out of 6 technical replicates). Out of the top 20, 7 glycans contain α2,6 SA linkages and 13 contains α2,3 SA.
The effect of these HA mutations on virus replication was determined in Madin-Darby Canine Kidney (MDCK) which express both α2,3 and α2,6 sialic acid (40). Virus replication was investigated at 32°C and 37°C which represent the temperature gradient of the human respiratory tract and allows for differentiation of LAIV temperature dependent changes in replication (6, 8, 41–43). The WT HA and EA HA LAIV showed no differences in the kinetics of infectious virus production or in peak virus production at 32°C. However, the 2,3 EA HA LAIV replicated poorly at this temperature, suggesting that some aspect of HA function in MDCK cell cultures is impacted by the additions of I226Q S228G (Figure 3A). At 37°C, the WT HA LAIV produced the highest amount of infectious virus compared to the EA HA LAIV and the 2,3 EA HA LAIV (Figure 3B), indicating a temperature-dependent effect of these mutations on LAIV replication. Plaque appearance, morphology and size was then assessed using MDCK cells at 32°C. All three LAIVs produced clear, distinct plaques (Figure 3C). EA HA LAIV plaque size was similar to the size of WT HA LAIV, however, the 2,3 EA HA LAIV produced smaller plaques compared to the WT HA LAIV (Figure 3D).
Figure 3: Replication of recombinant H3N2 LAIV viruses in MDCK cell cultures.
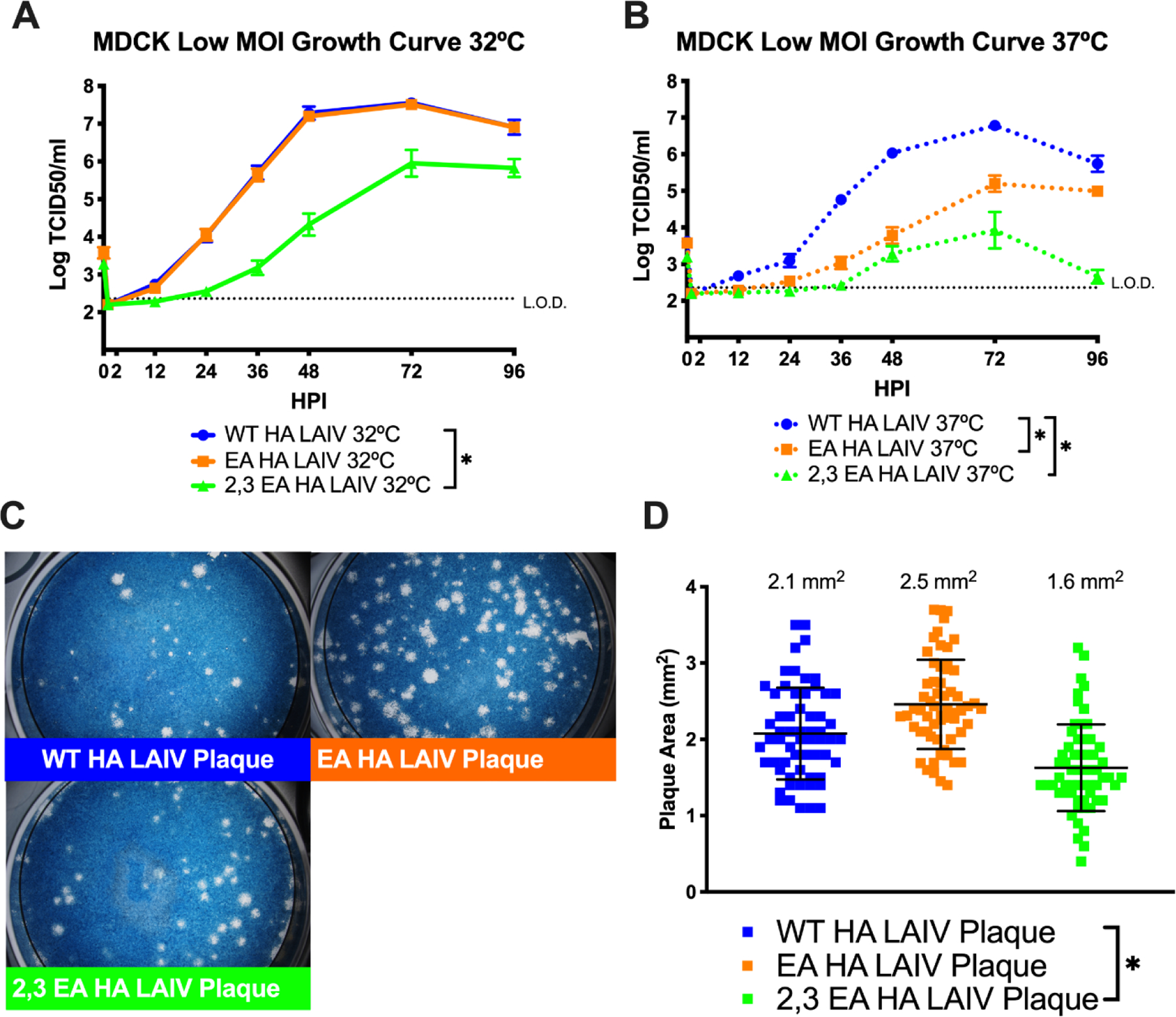
Low MOI growth curves with MDCK cells at 32°C (A) or 37°C (B) with the indicated A/Victoria/361/2011 LAIV recombinant viruses. Hours post infection (HPI) on X axis, log10 of TCID50/ml on Y axis. Data are pooled from 3 independent experiments with four replicates per virus per experiment (total n = 12 wells per virus timepoint). Data were analyzed with *p<.05 and two-way repeated measures ANOVA with Bonferroni multiple comparison posttest. The limit of detection (L.O.D.) is indicated with a dotted line at log 2.37 TCID50/ml. (C) Plaque assay performed with recombinant LAIV expressing WT, EA or 2,3 EA HA proteins at 32°C. (D) Quantification of plaque area from 30–50 individual plaques per virus from 3 independent experiments. *p<.05 unpaired T test. Error bars in (A) and (B) are SEM, error bars in (D) are SD.
Virus replication was then tested on primary hNEC cultures, which have previously been shown to be a faithful model of the human nasal epithelium and allow for better penetrance of virus replication phenotypes (6, 8, 24). The WT HA LAIV yielded significantly higher amounts of infectious virus compared to both the EA HA LAIV and the 2,3 EA HA LAIV at both 32°C (Figure 4A) and 37°C (Figure 4B), indicating LAIV binding to α2,3 sialic acid results in less efficient virus replication in hNEC cultures. To determine the effects of egg adapted mutations on epithelial cell antiviral responses, basolateral supernatant from hNEC cultures was tested for the presence of interferon lambda I/II/III (IFN-λ), a factor known to be produced by hNEC cultures during infection (13). The WT HA LAIV induced the highest amount of IFN-λ at 32ªC (Figure 4C) and 37°C (Figure 4D) compared to the EA HA LAIV and 2,3 EA HA LAIV which was directly correlated with the degree of virus replication (Figure 4A and 4B).
Figure 4: Replication of recombinant H3N2 LAIV viruses in hNEC cultures.
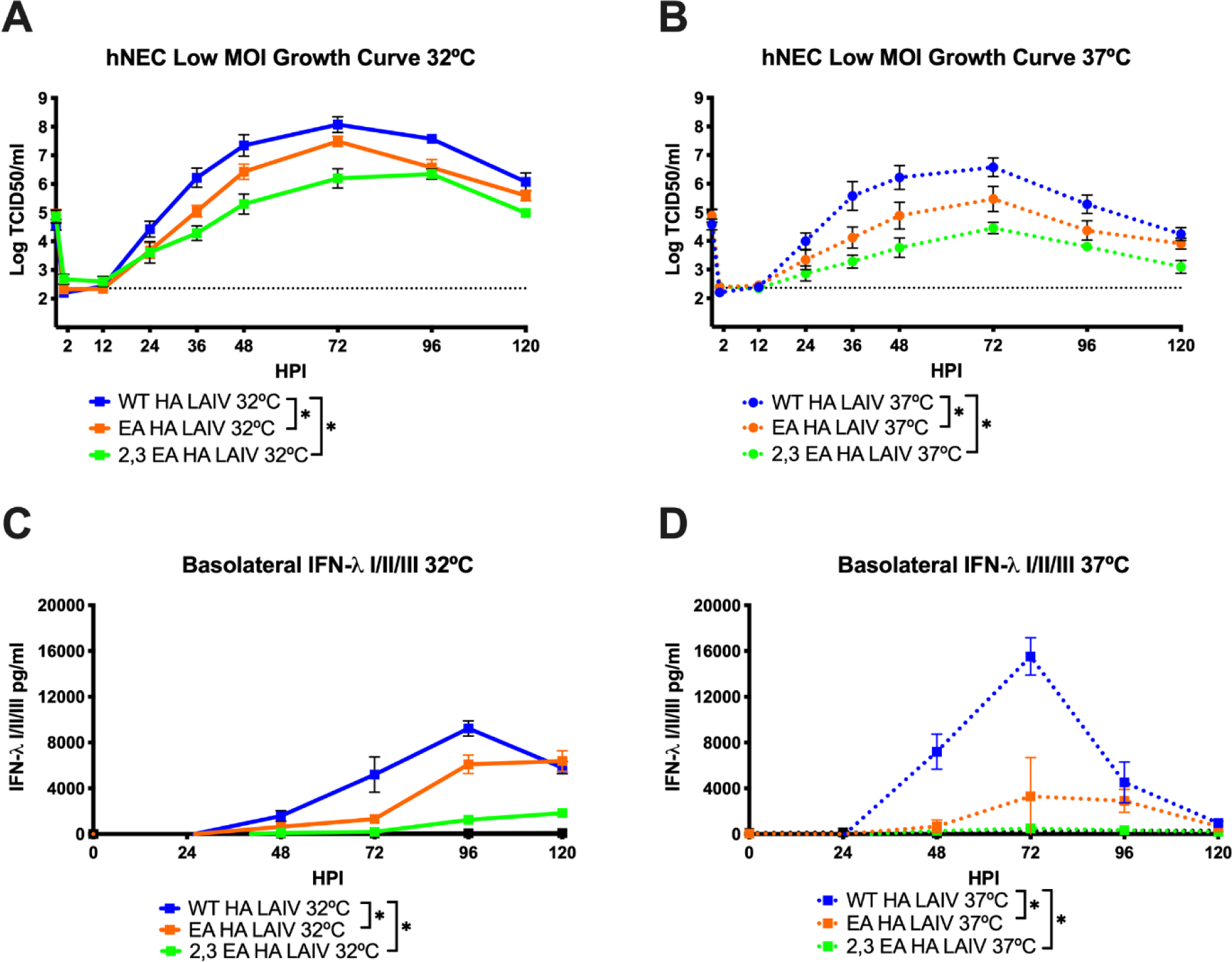
Low MOI growth curves with hNECs at 32°C (A) or 37°C (B) with the indicated A/Victoria/361/2011 LAIV recombinant viruses. Hours post infection (HPI) on X axis, Log of TCID50/ml on Y axis. Data are pooled from 2 independent experiments with four replicates per virus per experiment (total n = 8 wells per virus timepoint). The limit of detection (L.O.D.) is indicated with a dotted line at log 2.37 TCID50/ml. Quantification of IFN-λ I/II/III at 32°C (C) and 37°C (D) secreted in the basolateral media of hNEC cultures. Y axis pg/ml, X axis hours post infection (HPI). All data were analyzed with *p<.05 and two-way repeated measures ANOVA with Bonferroni multiple comparison posttest. Error bars are SEM.
Human nasal epithelial cell cultures contain ciliated epithelial cells, mucus secreting goblet cells, basal cells and other epithelial cell types (25, 44–49). Ciliated cells tend to express more α2,6 sialic acid than α2,3 sialic acid, and other cells in hNEC cultures tend to express more α2,3 sialic acid than α2,6 sialic acid (50, 51). We assessed how epithelial cell tropism was affected by HA receptor binding changes using flow cytometry. Human nasal epithelial cell cultures were infected at a high MOI of 3 for 14 hours, dissociated and immunostained for markers of cell lineage (Beta Tubulin IV, ciliated cells) and infection (H3 anti-serum) to determine the percentage of infection in all epithelial cells as well as the ciliated and non-ciliated epithelial cell subsets (Figure 5A). The gating strategy is shown with one example of each infection condition (Figure 5B). The WT HA LAIV infected a higher percentage of total epithelial cells compared to either the EA HA LAIV and 2,3 EA HA LAIV (Figure 5A). Additionally, the WT HA LAIV infected a higher percentage of ciliated cells (Beta Tubulin-IV +) compared to both the EA HA LAIV and 2,3 EA HA LAIV. When comparing infectivity of non-ciliated cells (Beta-Tubulin IV −), both the WT and EA HA LAIV infected a similar percentage, but the WT HA LAIV infected a higher percentage than the 2,3 EA LAIV. These data suggest that the increased preference for binding α2,6 sialic acid allows the WT HA LAIV to infect a higher percentage of ciliated epithelial cells, which tend to express more α2,6 sialic acid (50, 51). In contrast, the 2,3 EA HA LAIV showed significantly impaired infectivity when compared to WT HA LAIV for total, ciliated and non-ciliated cells (Figure 5A). This data, combined with the poor replication seen in MDCKs (Figure 3A–B) and hNECs (Figure 4A–B), suggest that changing the HA receptor binding preferences can lead to changes in infectious virus production, IFN-λ induction and epithelial cell tropism. It is also possible that this combination of amino acid changes on the 2,3 EA HA protein created an unstable or poorly functioning HA protein which contributed to an overall decrease of activity.
Figure 5: Tropism of LAIV in hNEC Cultures.
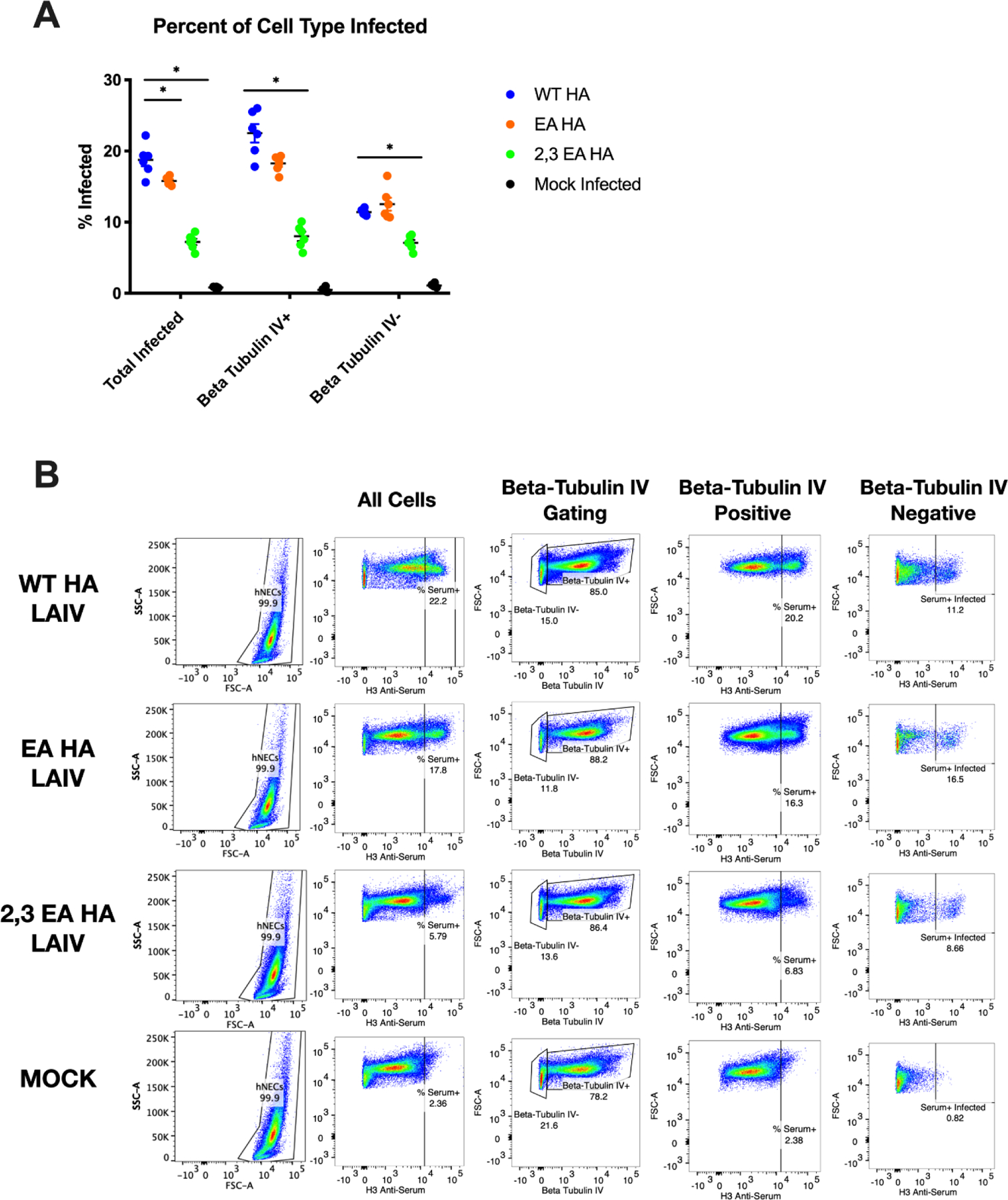
hNEC infected with a high MOI of three and virus tropism was determined via flow cytometry. Quantification of infection of total cells, Beta-Tubulin IV+ (ciliated epithelial cells) and Beta-Tubulin IV- (everything else in hNEC culture) (A). Gating strategy is shown for identifying lineages within heterogenous cell culture (B). Data are pooled from 3 independent experiments with 2 replicates per virus per experiment (total n = 6 hNEC wells tropism analysis). Data were analyzed with *p<.05 unpaired T test in A. Data was compared from WT HA LAIV percent infected to either EA or 2,3 EA HA. Mock is shown for background fluorescence in (A) and (B). Error bars in (A) are SEM. Acquisition of data on BD LSR II flow cytometer and analysis done with FlowJo 10.5.3 software and GraphPad Prism8.
Discussion
With the recent issues of poorly performing LAIV vaccine in the United States (52), this study was designed to understand how egg adaptation and the associated HA receptor amino acid mutations impact receptor specificity, viral fitness, innate immune induction and cellular tropism of an H3N2 LAIV vaccine strain. We demonstrated that egg adaptation of the HA protein can adversely affect LAIV replication on specific cell types and in a temperature dependent manner using recombinant viruses.
Using three different HA proteins with altered receptor binding specificity (Figure 1), we tested the hypothesis that egg adaptation and altering receptor preference would decrease LAIV replication. Human seasonal H3N2 WT viruses typically bind to α2,6 sialic acid (50, 51, 53–56). There is still an appreciable α2,3 sialic acid binding of the WT HA used in this study, as many human viruses bind both α2,6 and α2,3 sialic acid (57, 58). As expected, the EA HA receptor increased binding to the α2,3 sialic acid glycans with the 2,3 EA showing a further increased recognition of α2,3 sialic acid glycans (Figure 2). These results were expected and support previous glycan array studies using A/Victoria/361/2011 (17).
To test how receptor preferences dictated virus fitness we then analyzed virus growth. On immortalized MDCK cells, both the WT and EA virus produced a similar amount of infectious virus in a low MOI growth curve at 32°C, and both were significantly better than the 2,3 EA HA LAIV (Figure 3). At 37°C, the WT HA LAIV produced the highest amount of infectious virus titer compared to the EA HA LAIV and 2,3 EA HA LAIV, suggesting that HA egg associated amino acid changes were contributing to the attenuation phenotype of LAIV at 37°C. In our hNEC studies we showed that egg adaptation of the A/Victoria/361/2011 HA decreased replication at both 32°C and 37°C (temperature of the upper and lower respiratory tract respectively) as well as significantly decreased IFN-λ production at both 32°C and 37°C (Figure 4). Finally, we tested whether or not these HA receptor preferences dictated infectivity of the different HA LAIVs. We found that the WT HA LAIV infected more cells than either the EA HA or 2,3 EA HA LAIV in hNEC cultures in addition to having a slightly higher preference for ciliated cells (Figure 5). These results suggest that the receptor changes that resulted in differential recognition of sialic acid (Figure 2) impact the ability of EA and 2,3 EA HA LAIV to infect hNEC cultures. A decreased ability to infect epithelial cells can, in part, explain the lack of infectious virus production of the EA and 2,3 EA HA LAIV in low MOI growth curves. It is not known which ligands present in the CFG synthetic glycan array 5.3 are physiologically relevant to infection of hNEC cultures. Taken together, these results suggest that egg adapting the A/Victoria/361/2011 HA of LAIV further attenuates the LAIV and could significantly decrease vaccine effectiveness.
While HA egg-adaption has been shown to result in changes in receptor binding that can lead to antigenic changes, this study clearly demonstrates that those same changes can impact LAIV replication and innate immune responses. Since replication of LAIV is required for initiating a strong adaptive immune response, reduced LAIV replication could be an important and underappreciated contributor to poor LAIV vaccine efficacy. Thus, the failure of the 2013–14 LAIV vaccine was likely driven by two factors related to the egg adaption of the H3 HA protein – it was antigenically mismatched with the HA from circulating viruses and it lead to reduced LAIV replication. This does not mean that all egg-adaption mutations in H3 or H1 LAIVs will necessarily lead to reduced virus replication as it is possible that some egg adaption mutations may have a neutral effect on LAIV replication in primary respiratory epithelial cell cultures.
The reduced replication of EA HA and 2,3 EA HA LAIV also resulted in a reduction in the amount of IFNλ produced by infected hNEC cultures. Epithelial cells produce several factors that help recruit and activate immune cells, leading to efficient induction of both innate and adaptive immune responses (11, 13). Reduced LAIV replication may be contributing to reduced vaccine efficacy in two ways related to stimulating the adaptive immune response. First, the production of fewer infectious particles leads to a smaller amount of antigen and second, a weaker epithelial cell innate immune response could reduce the recruitment and activation of immune cells to the site of LAIV replication.
The contribution of physiologically relevant temperature ranges to LAIV replication and epithelial cell innate immune responses needs to be investigated more thoroughly. LAIV attenuation has often been studied at temperatures closer to 39°C, since the temperature dependent attenuation penetrates strongly at those temperatures. It is clear that the HA and perhaps the NA protein encoded by LAIV can contribute to temperature sensitivity and capturing this effect in future LAIV strains may help predict vaccine efficacy. Of course, in vitro replication does not completely recapitulate all the factors that are needed to induce a strong, adaptive immune response. However, a failure of LAIV to replicate on hNEC cultures could certainly be used as information that should merit more careful assessment of LAIV replication and vaccine efficacy in human populations.
There has been an assumption that LAIV replication and attenuation is independent of the specific HA and NA proteins that are used to generate the desired vaccine strains. In 2016, the Advisory Committee on Immunization Practices (ACIP) recommended the LAIV not be used for the 2016–2017 season. This was due to an ineffective H1N1 component in the previous 2015–2016 season (52). The H1N1 component of LAIV was an antigenically similar but not identical strain to that used in the IIV. In 2015/16 IIV had much greater efficacy compared to LAIV suggesting that the efficacy issues in LAIV were not due to a major antigenic difference between the two vaccine formulations. The H1 HA of LAIV used for the 2015/16 season has demonstrated very poor replication competency in MDCKs and human cell lines, suggesting that some aspect of egg adaptation or combining that particular H1 HA with the LAIV internal segments significantly decreased viral fitness (59). The in vitro replication properties are but one component that needs to be carefully considered when the HA and NA proteins are changed in the LAIV formulation. Egg adaption and specifically, the loss of glycosylation sites upon adaption has been a greater concern with H3 seasonal influenza vaccines compared to H1 seasonal influenza vaccines. Issues related to birth year and antigenic imprinting may impact the vaccine efficacy of both H1 and H3 viruses independently of in vitro replication. While the LAIV was recommended again starting in the 2018/19 season, a greater consideration of the effects of specific HA and NA proteins on LAIV replication would reduce the likelihood of future LAIV vaccine failures that are related to the ability of the vaccine to replicate in the upper respiratory tract. LAIV is still an attractive vaccine platform even with many potential pitfalls of replication competency. However, more focused research on stabilizing the HA protein during egg passaging or using an alternative substrate to grow vaccine stocks could solve the poor replication issues seen in certain LAIV vaccines.
Acknowledgements
We would like to thank the members of the Pekosz laboratory, the Sabra Klein laboratory, and the Kimberly Davis laboratory for data discussion and feedback. We thank Katherine Fenstermacher, Nick Wohlgemuth and Laura Canaday for their feedback and assistance with this project. The work was supported by Centers of Excellence in Influenza Research and Surveillance HHS N2772201400007C (AP) and T32 AI007417 (HP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.CDC. Disease Burden of Influenza 2020. [Available from: https://www.cdc.gov/flu/about/burden/index.html.
- 2.Cox NJ, Kitame F, Kendal AP, Maassab HF, Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2). Virology. 1988;167(2):554–67. [PubMed] [Google Scholar]
- 3.Toback SL, Levin MJ, Block SL, Belshe RB, Ambrose CS, Falloon J. Quadrivalent Ann Arbor strain live-attenuated influenza vaccine. Expert Rev Vaccines. 2012;11(11):1293–303. [DOI] [PubMed] [Google Scholar]
- 4.Sheldon EA, Jeanfreau R, Sliman JA, Charenkavanich S, Rousculp MD, Dubovsky F, et al. Immunogenicity of a quadrivalent Ann Arbor strain live attenuated influenza vaccine delivered using a blow-fill-seal device in adults: a randomized, active-controlled study*. Influenza Other Respir Viruses. 2013;7(6):1142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeneca A FluMist Quadrivalent 2020. [Available from: https://www.flumistquadrivalent.com/.
- 6.Fischer WA 2nd, King LS, Lane AP, Pekosz A. Restricted replication of the live attenuated influenza A virus vaccine during infection of primary differentiated human nasal epithelial cells. Vaccine. 2015;33(36):4495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez L, Blanco-Lobo P, Reilly EC, Maehigashi T, Nogales A, Smith A, et al. Comparative Study of the Temperature Sensitive, Cold Adapted and Attenuated Mutations Present in the Master Donor Viruses of the Two Commercial Human Live Attenuated Influenza Vaccines. Viruses. 2019;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wohlgemuth N, Ye Y, Fenstermacher KJ, Liu H, Lane AP, Pekosz A. The M2 protein of live, attenuated influenza vaccine encodes a mutation that reduces replication in human nasal epithelial cells. Vaccine. 2017;35(48 Pt B):6691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould PS, Easton AJ, Dimmock NJ. Live Attenuated Influenza Vaccine contains Substantial and Unexpected Amounts of Defective Viral Genomic RNA. Viruses. 2017;9(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Sobrido L, Peersen O, Nogales A. Temperature Sensitive Mutations in Influenza A Viral Ribonucleoprotein Complex Responsible for the Attenuation of the Live Attenuated Influenza Vaccine. Viruses. 2018;10(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer WA 2nd, Chason KD, Brighton M, Jaspers I. Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures. Vaccine. 2014;32(15):1761–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanthier PA, Huston GE, Moquin A, Eaton SM, Szaba FM, Kummer LW, et al. Live attenuated influenza vaccine (LAIV) impacts innate and adaptive immune responses. Vaccine. 2011;29(44):7849–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forero A, Fenstermacher K, Wohlgemuth N, Nishida A, Carter V, Smith EA, et al. Evaluation of the innate immune responses to influenza and live-attenuated influenza vaccine infection in primary differentiated human nasal epithelial cells. Vaccine. 2017;35(45):6112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shcherbik S, Pearce N, Kiseleva I, Larionova N, Rudenko L, Xu X, et al. Implementation of new approaches for generating conventional reassortants for live attenuated influenza vaccine based on Russian master donor viruses. J Virol Methods. 2016;227:33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiseleva IV, Isakova IN, Larionova NV, Oleinik ES, Rudenko LG. [Efficacy of production of reassortant of epidemic strains and cold-adapted influenza viruses in chicken embryo and MDCK cells]. Zh Mikrobiol Epidemiol Immunobiol. 2007(6):40–5. [PubMed] [Google Scholar]
- 16.Gambaryan AS, Robertson JS, Matrosovich MN. Effects of egg-adaptation on the receptor-binding properties of human influenza A and B viruses. Virology. 1999;258(2):232–9. [DOI] [PubMed] [Google Scholar]
- 17.Parker L, Wharton SA, Martin SR, Cross K, Lin Y, Liu Y, et al. Effects of egg-adaptation on receptor-binding and antigenic properties of recent influenza A (H3N2) vaccine viruses. J Gen Virol. 2016;97(6):1333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambaryan AS, Tuzikov AB, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6’-sialyl(N-acetyllactosamine). Virology. 1997;232(2):345–50. [DOI] [PubMed] [Google Scholar]
- 19.Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, et al. Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One. 2014;9(3):e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobey S, Gouma S, Parkhouse K, Chambers BS, Ertl HC, Schmader KE, et al. Poor Immunogenicity, Not Vaccine Strain Egg Adaptation, May Explain the Low H3N2 Influenza Vaccine Effectiveness in 2012–2013. Clin Infect Dis. 2018;67(3):327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shcherbik S, Pearce N, Carney P, Bazhenova E, Larionova N, Kiseleva I, et al. Evaluation of A(H1N1)pdm09 LAIV vaccine candidates stability and replication efficiency in primary human nasal epithelial cells. Vaccine X. 2019;2:100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hawksworth A, Lockhart R, Crowe J, Maeso R, Ritter L, Dibben O, et al. Replication of live attenuated influenza vaccine viruses in human nasal epithelial cells is associated with H1N1 vaccine effectiveness. Vaccine. 2020;38(26):4209–18. [DOI] [PubMed] [Google Scholar]
- 23.Parker L, Ritter L, Wu W, Maeso R, Bright H, Dibben O. Haemagglutinin stability was not the primary cause of the reduced effectiveness of live attenuated influenza vaccine against A/H1N1pdm09 viruses in the 2013–2014 and 2015–2016 seasons. Vaccine. 2019;37(32):4543–50. [DOI] [PubMed] [Google Scholar]
- 24.Wohlgemuth N, Lane AP, Pekosz A. Influenza A Virus M2 Protein Apical Targeting Is Required for Efficient Virus Replication. J Virol. 2018;92(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramanathan M Jr., Lane AP. Innate immunity of the sinonasal cavity and its role in chronic rhinosinusitis. Otolaryngol Head Neck Surg. 2007;136(3):348–56. [DOI] [PubMed] [Google Scholar]
- 26.WHO. Recommended composition of influenza virus vaccines for use in the 2013–14 northern hemisphere influenza season 2013. [Available from: https://www.who.int/influenza/vaccines/virus/recommendations/2013_14_north/en/.
- 27.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96(16):9345–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73(11):9679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, et al. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol. 2006;80(15):7469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrd-Leotis L, Cummings RD, Steinhauer DA. The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase. Int J Mol Sci. 2017;18(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrd-Leotis L, Gao C, Jia N, Mehta AY, Trost J, Cummings SF, et al. Antigenic Pressure on H3N2 Influenza Virus Drift Strains Imposes Constraints on Binding to Sialylated Receptors but Not Phosphorylated Glycans. J Virol. 2019;93(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss G, Chen X, Brindley MA, Campbell P, Afonso CL, Ke Z, et al. Capturing enveloped viruses on affinity grids for downstream cryo-electron microscopy applications. Microsc Microanal. 2014;20(1):164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McQuillan AM, Byrd-Leotis L, Heimburg-Molinaro J, Cummings RD. Natural and Synthetic Sialylated Glycan Microarrays and Their Applications. Front Mol Biosci. 2019;6:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glycomics CfF. MAMMALIAN PRINTED ARRAY VERSION 5.2 2020. [Available from: http://www.functionalglycomics.org/static/consortium/resources/resourcecoreh8.shtml.
- 35.Chan MCW, Wang MH, Chen Z, Hui DSC, Kwok AK, Yeung ACM, et al. Frequent Genetic Mismatch between Vaccine Strains and Circulating Seasonal Influenza Viruses, Hong Kong, China, 1996–2012. Emerg Infect Dis. 2018;24(10):1825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens J, Chen LM, Carney PJ, Garten R, Foust A, Le J, et al. Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J Virol. 2010;84(16):8287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mochalova L, Gambaryan A, Romanova J, Tuzikov A, Chinarev A, Katinger D, et al. Receptor-binding properties of modern human influenza viruses primarily isolated in Vero and MDCK cells and chicken embryonated eggs. Virology. 2003;313(2):473–80. [DOI] [PubMed] [Google Scholar]
- 38.Takemae N, Ruttanapumma R, Parchariyanon S, Yoneyama S, Hayashi T, Hiramatsu H, et al. Alterations in receptor-binding properties of swine influenza viruses of the H1 subtype after isolation in embryonated chicken eggs. J Gen Virol. 2010;91(Pt 4):938–48. [DOI] [PubMed] [Google Scholar]
- 39.Pekosz A, Newby C, Bose PS, Lutz A. Sialic acid recognition is a key determinant of influenza A virus tropism in murine trachea epithelial cell cultures. Virology. 2009;386(1):61–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson SW, Lorbach JN, Nolting JM, Stull JW, Jackwood DJ, Davis IC, et al. Madin-Darby canine kidney cell sialic acid receptor modulation induced by culture medium conditions: Implications for the isolation of influenza A virus. Influenza Other Respir Viruses. 2019;13(6):593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindemann J, Leiacker R, Rettinger G, Keck T. Nasal mucosal temperature during respiration. Clin Otolaryngol Allied Sci. 2002;27(3):135–9. [DOI] [PubMed] [Google Scholar]
- 42.Laporte M, Stevaert A, Raeymaekers V, Boogaerts T, Nehlmeier I, Chiu W, et al. Hemagglutinin Cleavability, Acid Stability, and Temperature Dependence Optimize Influenza B Virus for Replication in Human Airways. J Virol. 2019;94(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McFadden ER Jr., Pichurko BM, Bowman HF, Ingenito E, Burns S, Dowling N, et al. Thermal mapping of the airways in humans. J Appl Physiol (1985). 1985;58(2):564–70. [DOI] [PubMed] [Google Scholar]
- 44.de Courcey F, Zholos AV, Atherton-Watson H, Williams MT, Canning P, Danahay HL, et al. Development of primary human nasal epithelial cell cultures for the study of cystic fibrosis pathophysiology. Am J Physiol Cell Physiol. 2012;303(11):C1173–9. [DOI] [PubMed] [Google Scholar]
- 45.Park DY, Kim S, Kim CH, Yoon JH, Kim HJ. Alternative Method for Primary Nasal Epithelial Cell Culture Using Intranasal Brushing and Feasibility for the Study of Epithelial Functions in Allergic Rhinitis. Allergy Asthma Immunol Res. 2016;8(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller L, Brighton LE, Carson JL, Fischer WA 2nd, Jaspers I. Culturing of human nasal epithelial cells at the air liquid interface. J Vis Exp. 2013(80). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Y, Zhao X, Yang Z. [Ciliogenesis in human nasal epithelial cells cultured at the air-liquid interface]. Lin Chuang Er Bi Yan Hou Ke Za Zhi. 2004;18(2):88–90. [PubMed] [Google Scholar]
- 48.Charles DD, Fisher JR, Hoskinson SM, Medina-Colorado AA, Shen YC, Chaaban MR, et al. Development of a Novel ex vivo Nasal Epithelial Cell Model Supporting Colonization With Human Nasal Microbiota. Front Cell Infect Microbiol. 2019;9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schagen J, Sly PD, Fantino E. Characterizing well-differentiated culture of primary human nasal epithelial cells for use in wound healing assays. Lab Invest. 2018;98(11):1478–86. [DOI] [PubMed] [Google Scholar]
- 50.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440(7083):435–6. [DOI] [PubMed] [Google Scholar]
- 51.Kumlin U, Olofsson S, Dimock K, Arnberg N. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir Viruses. 2008;2(5):147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CDC. ACIP votes down use of LAIV for 2016–2017 flu season 2016. [Available from: https://www.cdc.gov/media/releases/2016/s0622-laiv-flu.html.
- 53.Sassaki GL, Elli S, Rudd TR, Macchi E, Yates EA, Naggi A, et al. Human (alpha2-->6) and avian (alpha2-->3) sialylated receptors of influenza A virus show distinct conformations and dynamics in solution. Biochemistry. 2013;52(41):7217–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karakus U, Pohl MO, Stertz S. Breaking the Convention: Sialoglycan Variants, Coreceptors, and Alternative Receptors for Influenza A Virus Entry. J Virol. 2020;94(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008;16(4):149–57. [DOI] [PubMed] [Google Scholar]
- 56.Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012;2(2):160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nobusawa E, Ishihara H, Morishita T, Sato K, Nakajima K. Change in receptor-binding specificity of recent human influenza A viruses (H3N2): a single amino acid change in hemagglutinin altered its recognition of sialyloligosaccharides. Virology. 2000;278(2):587–96. [DOI] [PubMed] [Google Scholar]
- 58.Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.AstraZeneca. Update: Live Attenuated Influenza Vaccine (LAIV) 2017. [Available from: https://www.izsummitpartners.org/content/uploads/2017/05/10e-1_Bandell_Update-on-LAIV.pdf.
- 60.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, et al. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology. 1997;233(1):224–34. [DOI] [PubMed] [Google Scholar]
- 61.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74(18):8502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. [DOI] [PubMed] [Google Scholar]
- 63.Matrosovich MN, Gambaryan AS, Tuzikov AB, Byramova NE, Mochalova LV, Golbraikh AA, et al. Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology. 1993;196(1):111–21. [DOI] [PubMed] [Google Scholar]
- 64.Bradley KC, Galloway SE, Lasanajak Y, Song X, Heimburg-Molinaro J, Yu H, et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J Virol. 2011;85(23):12387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]


