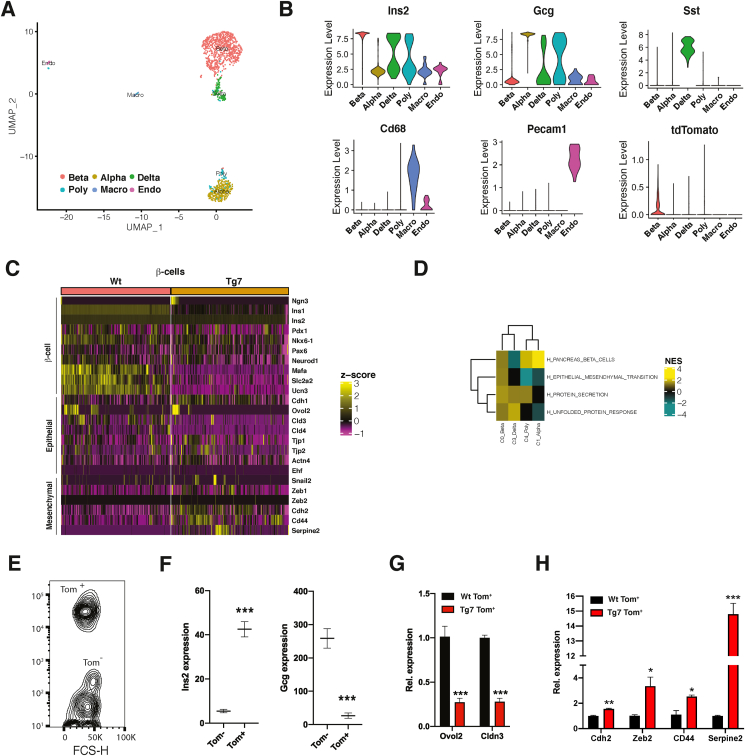
Figure 4.
scRNA-seq uncovers an EMT gene signature within β-cells of diabetic Tg7 mice. (A) Uniform manifold approximation and projection (UMAP) visualisation of islets cells from 12w Wt and Tg7 mice as a merged dataset with each dot consisting of a single cell. (B) Expression patterns of endocrine cell markers lns2, Gcg, and Sst defining α-, β-, and δ-cells, respectively. Polyhormonal (Poly) cells are defined as cells displaying co-expressing Ins2, Gcg, and Sst. Macrophage (Macro) and endothelial (Endo) cells are defined by the expression of Cd68 and Peacam1, respectively (C) Heatmap displaying differentially expressed genes in endocrine cell clusters from Wt and Tg7 mice. Magenta is low, black is intermediate, and yellow is high expression. (D) GSEA analysis of differentially expressed genes in endocrine islet cells of Tg7 mice. A selection of Hallmark gene sets is presented; FDR <0.25 (E) Contour plot of FACS-sorted tdTomato+ (Tom+) and tdTomato- (Tom−) cells from dissociated islets of Wtβ−Tom mice (F) Expression of Ins2 and Gcg in Tom+ and Tom− cells analysed by qPCR. Data related to data presented in panel E. (G) Expression of epithelial and mesenchymal markers in Wt Tom+ and Tg7 Tom+ cells obtained by FACS analysed by qPCR (n = 3). Unpaired Student's t-test. Data are means ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.