FIGURE 4.
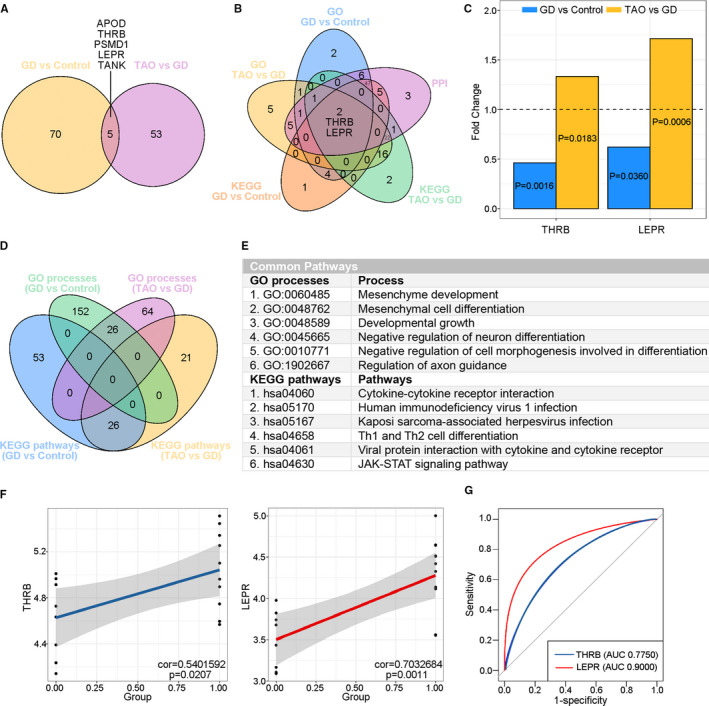
Identification of key immune‐related genes (IRGs) in TAO. (A) Venn diagram showed 75 and 58 differentially expressed IRGs in the GD vs HC set and TAO vs GD set, respectively. A total of 5 shared key IRGs were identified. (B) Five‐set Venn diagram showed a combination of all differentially expressed IRGs of Gene Ontology (GO) processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in the GD vs HC set and TAO vs GD set, respectively. A total of 2 common IRGs were identified. (C) Fold change and P value of the 2 common IRGs in the GD vs HC set and TAO vs GD set. (D) Four‐way Venn diagram of GO processes and KEGG pathways detected in the GD vs HC set and TAO vs GD set. A total of 26 GO processes and 6 KEGG pathways were identified in common between the GD vs HC set and TAO vs GD set. (E) Details of the common pathways from Venn diagram analysis. (F) The correlations of the expression of the 2 common key IRGs with TAO state in TAO vs GD set by linear regression analysis. (G) ROC curves of the 2 common key IRGs for diagnosing TAO. ROC curves were considered significant with AUC value >0.8. GD, Graves' disease; HC, healthy control; TAO, thyroid‐associated ophthalmopathy; AUC, area under curve
