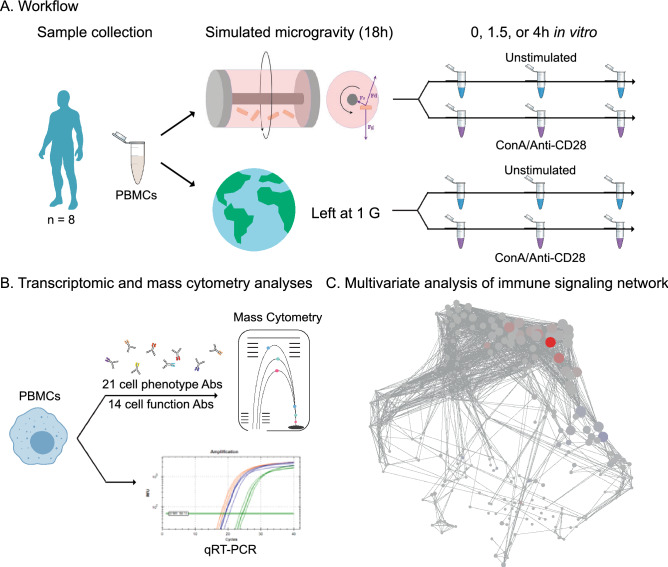
Figure 1.
Experimental workflow. (A) Blood samples from eight healthy volunteers were collected. Peripheral blood mononuclear cells (PBMCs) were isolated and exposed to simulated microgravity (sµG) or (1G) (static control) using a random positioning machine for 18 h, then stimulated with ConA and anti-CD28 (ConA/anti-CD28) or left unstimulated. (B) The activation and intracellular signaling responses of all major immune cell subsets were quantified using single-cell mass cytometry. In parallel, the mRNA expression of a select number of genes was quantified using qRT-PCR. (C) The high-dimensional immunological dataset was visualized as an immune signaling correlation network. Cell-type-specific immune responses that differed between the sµG and 1G conditions were identified using a multivariate Elastic Net (EN) method.