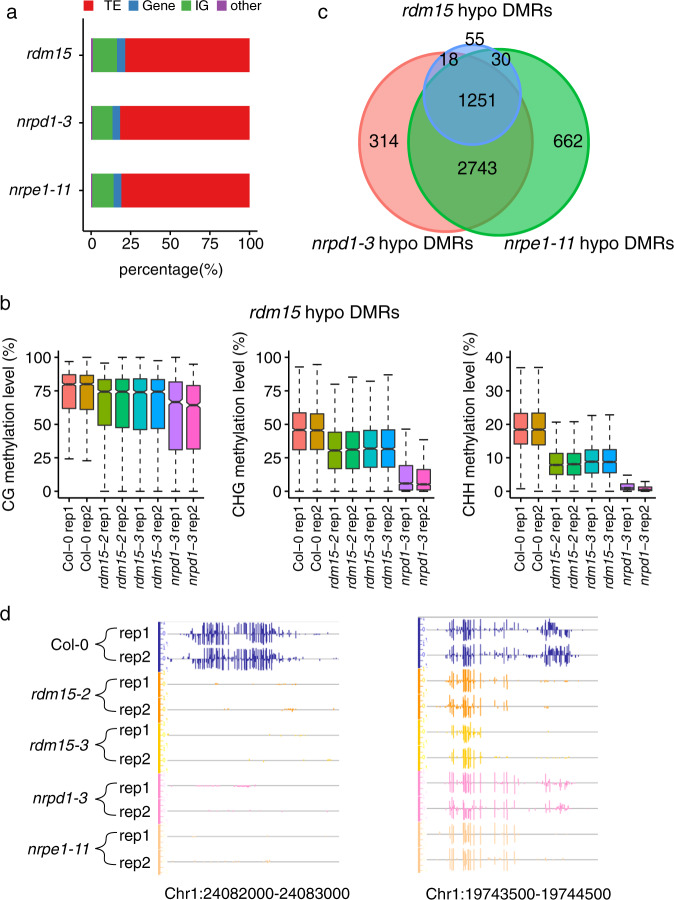
Fig. 2. RDM15 is required for RdDM-dependent DNA methylation.
a Compositions of DMRs among TEs, genes, and intergenic regions (IGs). DMRs (relative to the WT) were identified in rdm15, nrpd1-3, and nrpe1-11. b Box plots showing CG, CHG, and CHH methylation levels of rdm15 DMRs in Col-0, rdm15-2, rdm15-3, and nrpd1-3. Data from two replicates are shown. c Venn diagram showing the numbers of hypomethylated DMRs that overlapped between rdm15, nrpd1-3, and nrpe1-11. d The Integrative Genome Browser (IGB) display of whole-genome bisulfite sequencing (WGBS) data is shown for several representative rdm15 DMRs in Col-0, rdm15-2, rdm15-3, and nrpd1-3. Vertical bars on each track indicate DNA methylation levels. The coordinates of DMRs are indicated below the snapshots. Two replicates are shown for each genotype.