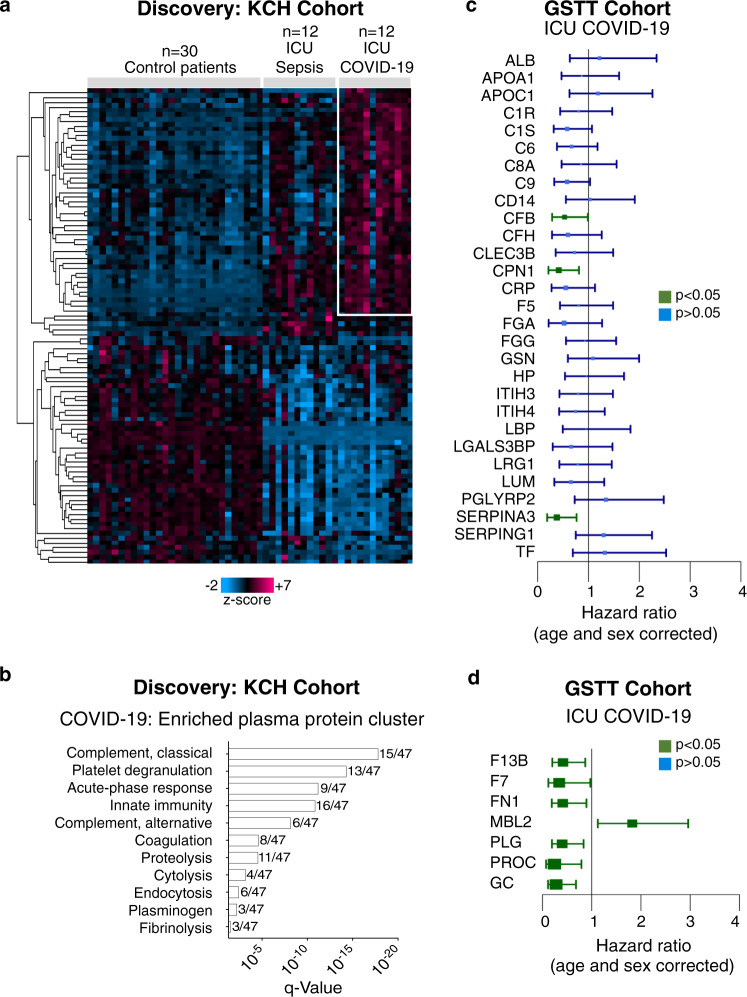
Fig. 3. COVID-19 circulating proteome signature and associations with 28-day mortality.
a Plasma proteome profiling was conducted using a data-independent acquisition–mass spectrometry (DIA-MS) approach with spiked standards for 500 proteins. Hierarchical cluster analysis was conducted upon significantly changing plasma proteins across control patients before elective cardiac surgery (n = 30), ICU patients with sepsis (n = 12), and ICU patients with COVID-19 (n = 12, KCH). The heatmap highlights 47 proteins enriched in COVID-19. Kruskal–Wallis, Benjamini–Hochberg correction q < 0.05. b Gene ontology enrichment analysis was conducted upon these 47 proteins and significantly enriched pathways are represented. c Twenty-nine common proteins cross-referenced against two published proteomic studies, exploring protein markers of COVID-19 severity. The ability of these 29 proteins to predict 28-day mortality was explored in an independent ICU patient cohort (n = 62 patients, GSTT) by DIA–MS, and hazard ratios with 95% CI are shown. d Proteomic analysis by DIA-MS conducted upon the serum samples of the GSTT COVID-19 ICU cohort returned additional candidates that predict 28-day mortality as shown on hazard ratio plots with 95% CI (n = 62 patients, GSTT). Significance was determined through the Mann–Whitney U test, correcting for age and sex and applying the Benjamini-Hochberg procedure. All statistical analyses are two-tailed.