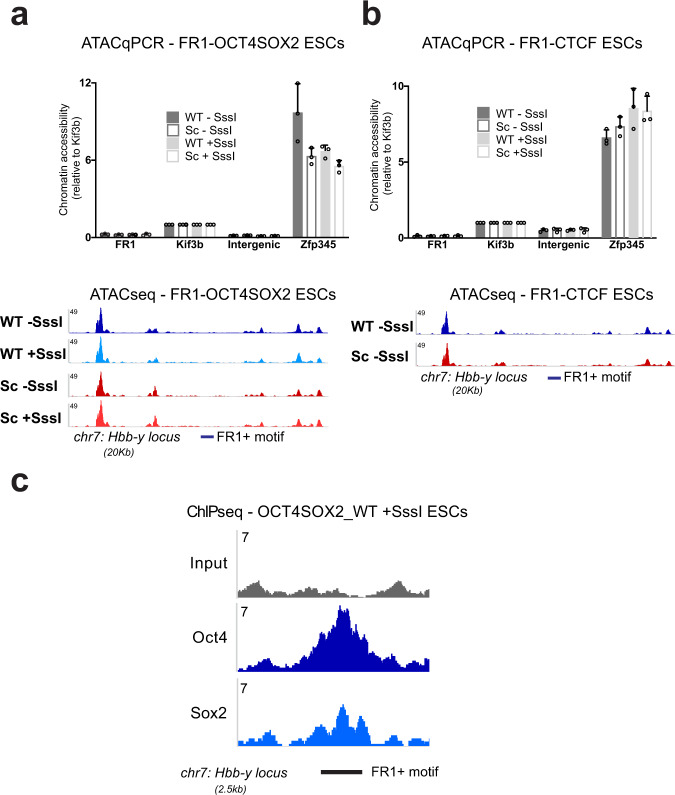
Fig. 5. SPF activity is not sufficient, in isolation, to generate chromatin accessibility.
a, b Upper panels. Results of ATAC-qPCR experiments in FR1_OCT4SOX2 ESCs (a) and FR1_CTCF ESCs (b) containing WT and Sc motifs in −SssI and +SssI conditions. We measured very low levels of chromatin accessibility at the FR1 locus in all four conditions when compared to known accessible (Zfp345 large ATAC-peak; Kif3b medium ATAC-peak) and inaccessible (Intergenic small ATAC peak) regions of the chromatin in mESCs97. Results are shown as mean+SD of n = 3 biologically independent replicates. Source data are provided as a Source data file. Lower panels. Representative genome browser tracks of the chromatin accessibility landscape around the FR1 locus. ATAC-Seq experiments were performed in FR1-OCT4SOX2 ESCs containing WT/±SssI and Sc/±SssI motifs (a) and in FR1-CTCF ESCs containing WT/−SssI and Sc/−SssI motifs (b). The ATAC-Seq signal is low over the FR1 locus compared to neighbouring accessibility peaks, highlighting the low levels of chromatin accessibility, which remain unchanged despite SPF binding in the WT conditions. c OCT4 and SOX2 ChIP-Seq in FR1 OCT4SOX2 ESCs reveals binding of both TFs at the FR1 locus. Displayed are representative genome browser tracks of the ChIP-Seq data across the FR1 locus.