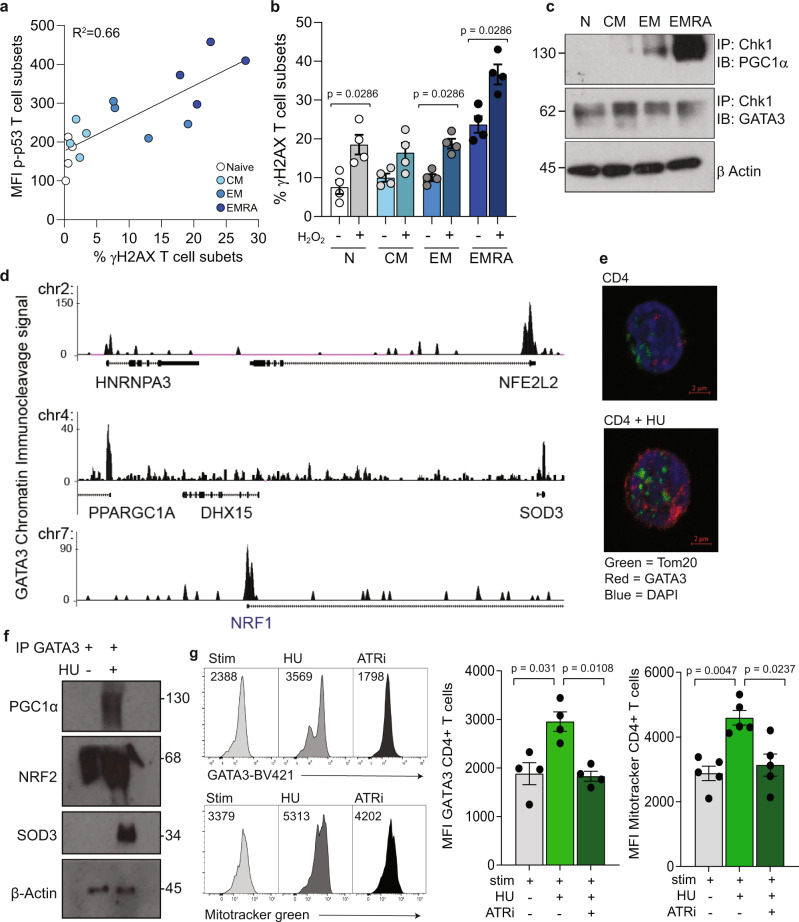
Fig. 3. DNA damage recruits a GATA3-PGC1α complex to induce mitochondria biogenesis in CD4+ EMRA T cells.
a Linear regression analysis of the DNA damage marker ƴH2AX and p-p53 in CD27/CD45RA defined CD4+ T cell subsets (n = 4 biologically independent samples). b DNA damage in CD4+ T-cell subsets following the induction of oxidative stress using H2O2 (n = 4 biologically independent samples). c Western blot showing Chk1 immunoprecipitation in the CD27/CD45RA defined CD4+ T cell subsets quantifying the presence of GATA3 and PGC1α bound to the Chk1 antibody (n = 3 biologically independent samples). d Chromatin immunocleavage showing GATA3 binding to the promoter regions of NRF1 (NRF1), NRF2 (NFE2L2), SOD3 (SOD3), and PGC1α (PPARGC1A) in naïve CD4+ T cells. e Tom20 and GATA3 staining in CD4+ T cells following treatment with 400 μM hydroxyurea (n = 3 biologically independent samples). f Western blot showing GATA3 immunoprecipitation in CD4+ T cells following hydroxyurea treatment quantifying the presence of PGC1α, NRF2 and SOD3 (n = 3 biologically independent samples). g Representative flow cytometry histograms and graph of GATA3 and mitochondrial mass measured using mitotracker green following treatment of CD4+ T cells with and without hydroxyurea and 1 μM AZD6738 (n = 4 biologically independent samples). p values were calculated using a two-way paired Mann–Whitney U test for part b and a Kruskal–Wallis test followed by Dunn multiple comparison for post hoc testing for part g. Graphs show SEM.