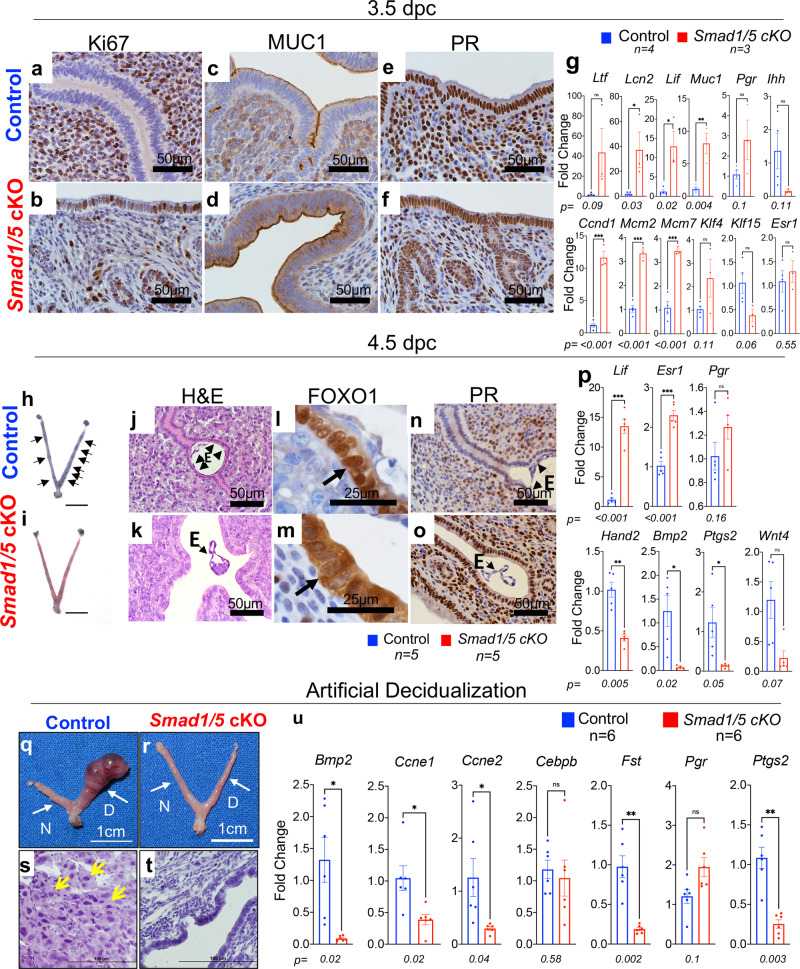
Fig. 3. Abnormal endometrial receptivity at 3.5 dpc and defective embryo implantation at 4.5 dpc in Smad1/5 cKO mice.
a–f Histological analysis of 3.5 dpc uteri from control (a, c, e) and Smad1/5 cKO (b, d, f) mice was performed by staining with Ki67 (a–b), MUC1 (c–d), or progesterone receptor (PR) (e–f). g qPCR analysis from the endometrial epithelium of control (blue bars) or Smad1/5 cKO (red bars) mice. Paired, two-tailed, t test, mean ± SEM, *p < 0.05, **p < 0.001, ***P < 0.0001. h–i Whole uteri of control (h) and Smad1/5 cKO (i) mice isolated at 4.5 dpc after injection with Chicago Sky Blue dye; implantation sites in the control mice can be visualized as blue bands (indicated by black arrows). Size bar = 1 cm. j–k H&E-stained cross-sections from 4.5 dpc control (j) and Smad1/5 cKO (k) uteri. e=embryo. l–o IHC of FOXO1 i–m and progesterone receptor (PR) n–o in 4.5 dpc uterine cross-sections of control (l–n) and Smad1/5 cKO (m–o) mice. Arrows in l, m indicate the nuclear staining of FOXO1 in the controls (l) and cytoplasmic FOXO1 staining in the Smad1/5 cKO mice (m) E embryo. Representative image of the embryo, observed in at least three specimens from different mice. p qPCR analysis of implantation-related markers in control (blue bars, n = 5) and Smad1/5 cKO (red bars, n = 5) uterine tissues collected at 4.5 dpc. Histology and qPCR analyses were performed in at least three samples of each genotype. Histograms represent mean ± SEM. Paired, two-tailed, t test, *p < 0.033, **p < 0.002, ***P < 0.001. q–u Analysis of decidualization reveals that compared with controls, Smad1/5 cKO cannot respond to the artificial induction of decidualization. Gross images of control (q) and Smad1/5 cKO (r) uteri, (D, decidual horn; N, non-decidual horn; indicated by white arrows). s–t H&E stains of uterine cross-sections of the decidual horns of control (s) and Smad1/5 cKO (t) mice. Yellow arrows in (s) indicate decidualized cells. u Gene expression analysis by qPCR of control (blue bars, n = 6) and Smad1/5 cKO (red bars, n = 6) uterine tissues that received the decidual stimulus. Histology and qPCR analyses were performed in at least three samples of each genotype. Histograms represent mean ± SEM. Paired, two-tailed, t test, *p < 0.033, **p < 0.002, ***P < 0.001.