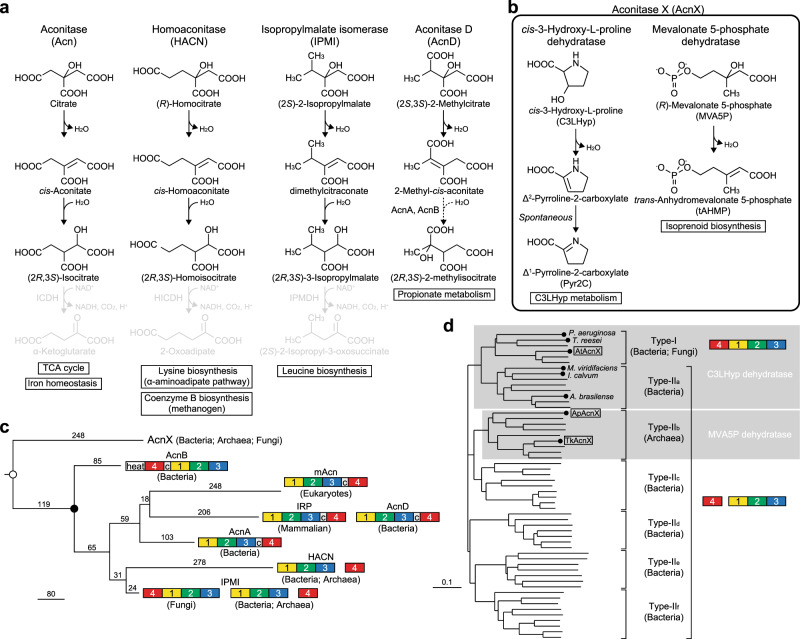
Fig. 1. Aconitase superfamily members.
Schematic reactions of four known functional members of the aconitase superfamily (a) and two functional members of the AcnX subfamily (b) . Each physiological role is in a box. In propionate metabolism, AcnD catalyzes only the dehydration of (2 S,3 S)-2-methylcitrate to 2-methyl-cis-acotinate, which is subsequently metabolized to (2 S,3 R)-2-methylisocitrate by either AcnA and AcnB. c Phylogenetic tree of the aconitase superfamily. A linear representation of the sequential domain arrangement of eight phylogenetic subfamilies is also included. “c” is the connector domain and “heat” is a protein–protein interaction domain found only in AcnB2. This phylogenetic tree was constructed based on the sequence identity of domain 4, in the National Center for Biotechnology Information (NCBI). The closed circle indicates the common ancestor for other aconitase enzymes that had previously been proposed, which contains the [4Fe-4S] cluster. The open circle indicates the common ancestor for the aconitase superfamily that is proposed in the present study. d Phylogenetic tree of the AcnX subfamily based on sequence similarity. The AcnX subfamily is further classified into AcnXType-I, consisting of a single polypeptide from bacteria and fungi, and AcnXType-II in a number of bacteria (AcnXType-IIa, AcnXType-IIc AcnXType-IIf) and archaea (AcnXType-IIb), which consists of (fragmented) small and large polypeptide chains. Among them, AcnXType-I and AcnXType-IIa, and AcnXType-IIb correspond to C3LHyp dehydratase and MVA5P dehydratase, respectively, in Fig. 1b. The circles at the end of each branch are the enzymes that have been functionally characterized. The large subunit of AcnXType-II was used for this phylogenetic analysis. A more-detailed comparison is shown in Supplementary Fig. 8.