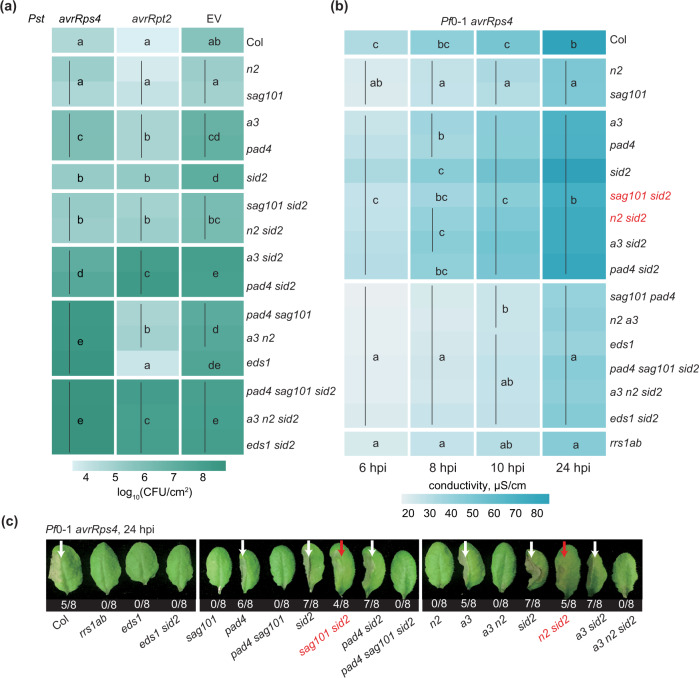
Fig. 2. Removal of the SAG101–NRG1 and ICS1 sectors reveals PAD4–ADR1-promoted TNL cell death.
a A heatmap representation of Pst avrRps4, Pst avrRpt2 or Pst (empty vector, EV) growth at 3 dpi in leaves of indicated genotypes (syringe infiltration, OD600 = 0.0005). The significance codes are based on Tukey’s HSD test (α = 0.001, n = 12 for Pst avrRps4 and Pst avrRpt2, n = 16 for Pst). Data points were combined from three (Pst avrRps4, Pst avrRpt2) or four (Pst) independent experiments, each with four replicates. Note: significance codes are assigned based on the statistical analysis per treatment and should be read columnwise. b A heatmap of quantitative cell death assays conducted on leaves of indicated genotypes after infiltration utilising the Pseudomonas fluorescens 0–1 effector tester strain (hereafter Pf0-1) delivering avrRps4 (OD600 = 0.2). Cell death was measured by electrolyte leakage from bacteria-infiltrated leaf discs at 6, 8, 10 and 24 hpi. Data are displayed as means from four experiments, each with four replicates (n = 16). Statistical significance codes for the difference in means are based on Tukey’s HSD test (α = 0.001). In mutants marked in red, the PAD4–ADR1s cell death branch operates in TNLRRS1-RPS4 immunity when SAG101–NRG1s and ICS1 pathways are not functional. Note: significance codes are assigned based on the statistical analysis per timepoint and should be read columnwise. Boxplot representation of the same data is provided in Supplementary Figs. 5 and 6. c Visual cell death symptoms at 24 hpi Pf0-1 avrRps4 infiltrating into leaf halves of indicated genotypes as in b. The ratio beneath each leaf indicates number of leaves with visible tissue collapse from all infiltrated leaves in two independent experiments. White arrows mark cell death visible as tissue collapse in a manner dependent on SAG101–NRG1s. Red arrows mark cell death in lines without functional SAG101–NRG1s and ICS1. Genotypes in red have functional PAD4–ADR1s but not SAG101–NRG1s. Pst Pseudomonas syringae pv. tomato DC3000, Pf0-1 effector tester strain of Pseudomonas fluorescens 0–1, CFU colony-forming units, EV empty vector.