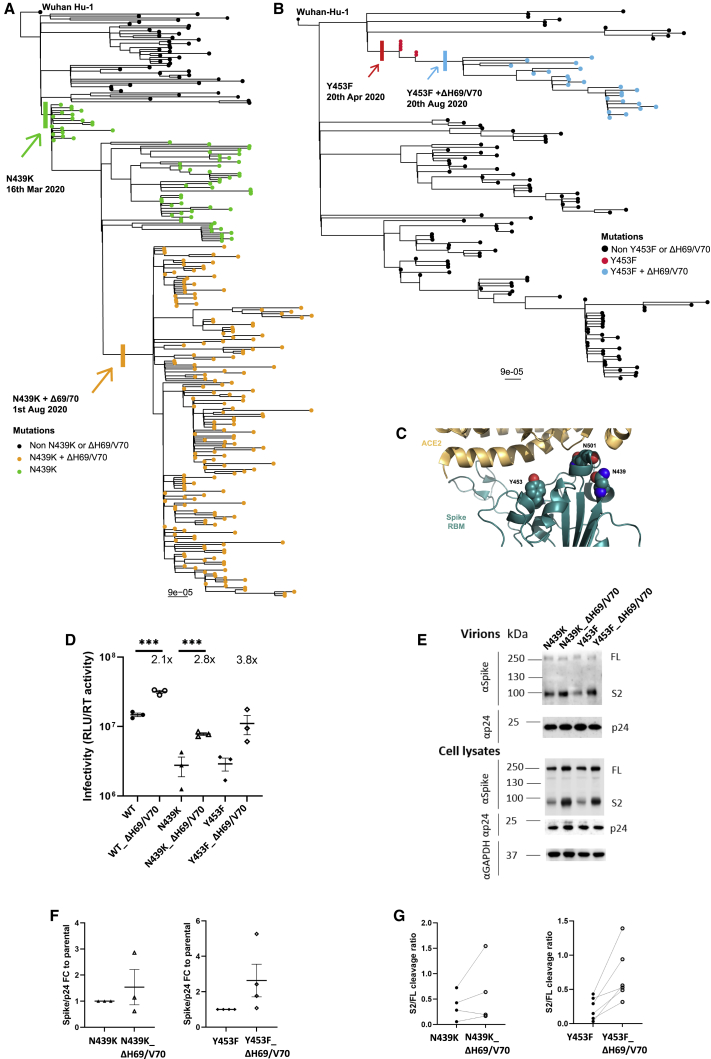
Figure 5.
ΔH69/V70 appears after spike N439K and Y453F and compensates for their reduced infectivity
(A and B) Maximum-likelihood phylogeny of global sequences carrying Spike mutant (A) N439K and (B) Y453F. All sequences in the GISAID database containing S:439K or S:Y453F (February 18, 2021) were downloaded, realigned to Wuhan-Hu-1 using MAFFT, and deduplicated.
(C) Representation of the Spike RBM:ACE2 interface (PDB: 6M0J) with residues N439, Y453, and N501, highlighted as spheres colored by element.
(D–F) Spike mutant ΔH69/V70 compensates for the infectivity defect of spike RBD mutations and is associated with increased spike incorporation into virions.
(D) Infectivity of spike (D614G) ΔH69/V70 in the absence and presence of spike RBD mutations. Shown is single-round infection by luciferase-expressing lentiviruses pseudotyped with SARS-CoV-2 spike protein on target HeLa cells stably transduced with ACE2. Mean and SEM are shown.
(E) Representative western blot of purified virions and cell lysates probed with antibodies against HIV-1 p24, SARS-CoV-2 spike S2, and GAPDH.
(F and G) Densitometric quantification of the (F) spike:p24 and (G) cleaved S2 spike:FL spike ratios for spike (D614G) ΔH69/V70 in the absence and presence of spike RBD mutations across multiple experiments in pelleted viruses. U, unit of reverse transcriptase (RT) activity.
Data are representative of at least two independent experiments. Student’s t test, ∗∗∗p < 0.001.