Summary
N6-methyladenosine (m6A) is emerging as a vital factor regulating neural differentiation. Here, we report that deficiency of Arhgef2, a novel cause of a neurodevelopmental disorder we identified recently, impairs neurogenesis, neurite outgrowth, and synaptic formation by regulating m6A methylation. Arhgef2 knockout decreases expression of Mettl14 and total m6A level significantly in the cerebral cortex. m6A sequencing reveals that loss of Arhgef2 reduces m6A methylation of 1,622 mRNAs, including Npdc1 and Cend1, which are both strongly associated with cell cycle exit and terminal neural differentiation. Arhgef2 deficiency decreases m6A methylations of the Npdc1 and Cend1 mRNAs via down-regulation of Mettl14, and thereby inhibits the translation of Npdc1 and nuclear export of Cend1 mRNAs. Overexpression of Mettl14, Npdc1, and Cend1 rescue the abnormal phenotypes in Arhgef2 knockout mice, respectively. Our study provides a critical insight into a mechanism by which defective Arhgef2 mediates m6A-tagged target mRNAs to impair neural differentiation.
Subject areas: Molecular neuroscience, Developmental neuroscience, Cellular neuroscience
Graphical abstract
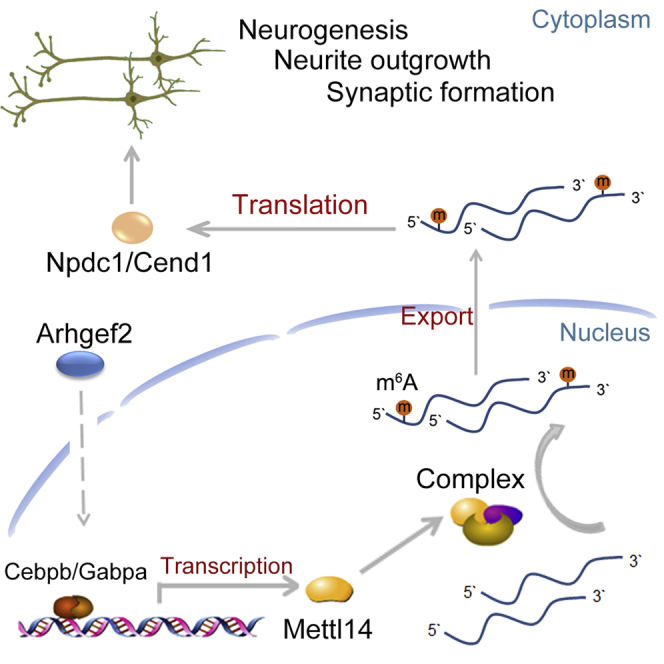
Highlights
-
•
Arhgef2 mediates total m6A level via Mettl14
-
•
Arhgef2 affects m6A methylations of the Npdc1 and Cend1 mRNAs
-
•
Decreased m6A methylations inhibits translation of Npdc1 and nuclear export of Cend1
-
•
Reduced protein expression of Npdc1 and Cend1 hinders neural differentiation
Molecular neuroscience; Developmental neuroscience; Cellular neuroscience
Introduction
ARHGEF2 (synonym GEF-H1, murine Lfc), a member of the Rho guanine nucleotide exchange factor family, is crucial for controlling the spatiotemporal activation of Rho GTPases (Ren et al., 1998; Krendel et al., 2002). It is now well established that ARHGEF2 controls microtubule dynamics through RhoA activation (Meiri et al., 2012; Birkenfeld et al., 2007; Tonami et al., 2011). Lack of ARHGEF2 is strongly associated with an alternation in the organization of the actin cytoskeleton at the cell edge (Nalbant et al., 2009). ARHGEF2 is required for mitotic spindle formation and orientation (Bakal et al., 2005; Yoshizaki et al., 2004). Recently, we successfully linked ARHGEF2 to neurologic disease by reporting a homozygous ARHGEF2 mutation in individuals with a neurodevelopmental phenotype including brain malformation (Ravindran et al., 2017). ARHGEF2 loss of function impaired neuronal migration by RhoA/ROCK/MLC pathway, which caused the abnormal location of precerebellar neurons in the hindbrain (Ravindran et al., 2017). Although we found that defective ARHGEF2 resulted in reduced neurogenesis of neocortical progenitors and probably contributed to microcephaly phenotype, little is known about whether ARHGEF2 regulates neurite outgrowth and synaptic formation during neural differentiation. Thus, the molecular mechanism that how ARHGEF2 mediates neural differentiation of brain cortex needs to be further investigated.
Dynamic methylation at the N6 site of adenosine (N6-methyladenosine, m6A) has been indicated as the most prevalent internal mRNA modification in eukaryotes (Zhuang et al., 2019). A growing body of evidence has revealed that 0.1%–0.4% of all adenosines in mammal transcripts are modified by m6A, which enriched in long exons, near transcription start sites, and stop codons (Meyer and Jaffrey, 2014; Wang and Zhao, 2016). It is involved in various biological processes, including mRNA splicing, localization, stability, and translation (Haussmann et al., 2016; Roundtree et al., 2017; Wang et al., 2014; Li et al., 2017). In recent years, m6A is emerging as a key factor regulating neural differentiation. Deficiency of methyltransferase-like Mettl14 leads to the reduced nuclear export of m6A-modified mRNAs regulating neural differentiation (Edens et al., 2019). Depletion of m6A by Mettl14 knockout in embryonic mouse brains prolongs the cell cycle of radial glia cells and extends cortical neurogenesis into postnatal stages (Yoon et al., 2017), phenocopying Arhgef2 knockout mice we constructed in this study. Furthermore, in view of drastically increased m6A levels in adult mouse brain compared with embryogenesis period (Meyer et al., 2012), it remains possible that m6A modification plays a unique role in the adult brain. Indeed, a compelling evidence points out that deletion of Mettl14 in the adult brain severely impairs learning and performance, which is correlated to protein synthesis in synapses of striatum (Koranda et al., 2018). However, the mechanism of m6A modification on neurite outgrowth and synaptic formation in postnatal brain cortex is still covered with a mysterious veil.
In this study, we cross-linked the function of Arhgef2 and modification of m6A regulated by Mettl14. The anomaly of neurogenesis, neurite outgrowth, and synaptic formation was demonstrated in an Arhgef2 knockout mouse model. We further illustrated the function of Mettl14 in this process and investigated underlying cellular and molecular mechanisms. Together, our data revealed a vital role of Arhgef2 in regulating neural differentiation by m6A-modified target mRNAs.
Results
Defective Arhgef2 caused microcephaly in adult mice
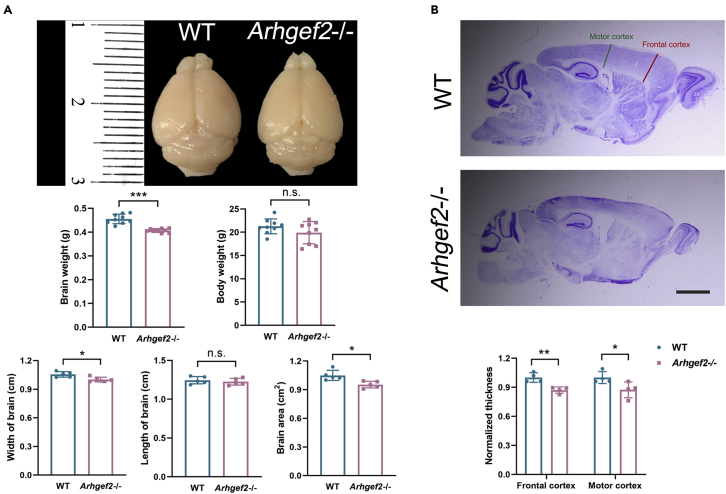
Previously, we reported a homozygous ARHGEF2 mutation in individuals with a neurodevelopmental phenotype including brain malformation (Ravindran et al., 2017). To explore the pathomechanism of brain malformation by loss of Arhgef2, we constructed an Arhgef2 knockout (Arhgef2−/−) mice model using CRISPR-Cas9 genome editing technology (Figure S1). A 1.14-fold decrease in brain weight was noted in response to Arhgef2 knockout, whereas the body weight had no significant change. Compared with wild-type littermates, the brain size was significantly decreased in Arhgef2−/− mice (Figure 1A). Nissl staining further exhibited reduced thickness of frontal and motor cortex in Arhgef2−/− mice (Figure 1B).
Figure 1.
Microcephaly in Arhgef2 knockout mouse model (postnatal 6 weeks)
(A) The brain weight and size were significantly decreased in Arhgef2−/− mice. n = 5–8 per genotype, ∗∗∗p < 0.001, ∗p < 0.05, n.s.: no significant difference, unpaired t test.
(B) Nissl staining of sagittal brain sections. Arhgef2−/− mice showed a reduced thickness of frontal and motor cortex region compared with wild-type ones, 30 μm thick; scale bar, 2.5 mm, n = 4 per genotype, ∗∗p < 0.01, ∗p < 0.05, unpaired t test. Data are represented as mean ± S.D.
Defective Arhgef2 led to abnormality of neural differentiation
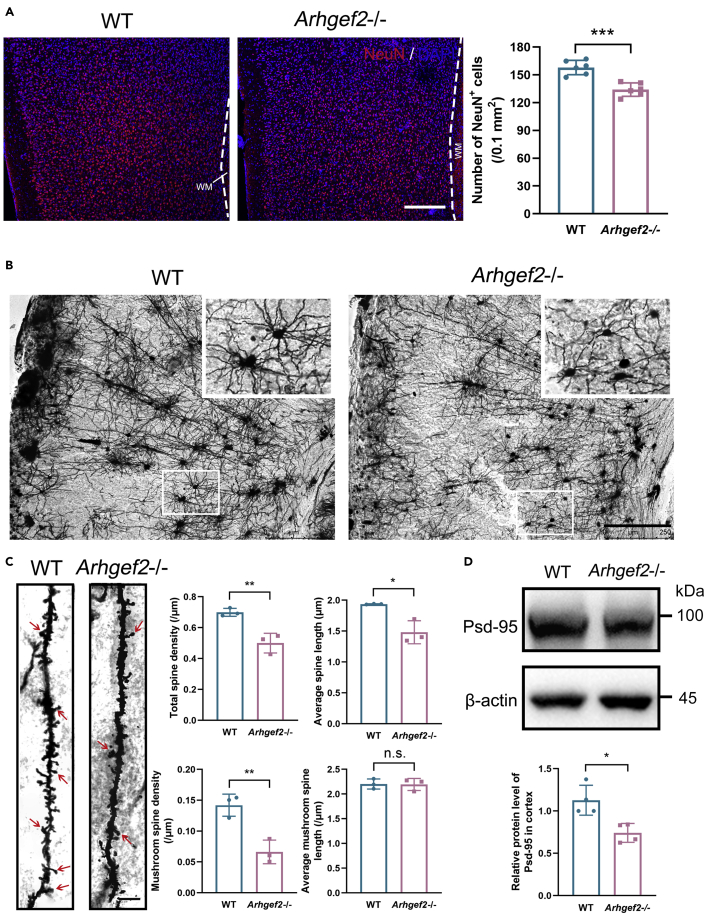
Our recent study indicated that, in mouse neocortex, Arhgef2 regulates neurogenesis from cortical precursor cells (Ravindran et al., 2017). As we expected, the number of NeuN+ cells significantly decreased in Arhgef2−/− mice brain cortex compared with wild-type mice (postnatal 8 weeks), and it showed no tendency at all layers (Figure 2A). To test whether the genetic removal of Arhgef2 affects neurite branching and synapse formation during neural differentiation, we examined the morphology of cortical neurites and dendrite spines in adult Arhgef2−/− mice. Golgi staining demonstrated that the spine density and length were significantly below the level of those of wild-type mice, with obviously abnormal morphology of neurites. Moreover, mature spines, which are characterized by mushroom-like shape, significantly reduced in Arhgef2−/− mice compared with wild-type mice (Figures 2B and 2C). We also analyzed the expression of presynaptic protein Synaptophysin and postsynaptic protein Psd-95. Compared with wild-type mice, expression of Psd-95 decreased in Arhgef2−/− mice (Figure 2D), whereas Synaptophysin showed no significant change (data not shown).
Figure 2.
Abnormality of neural differentiation in Arhgef2-knockout mice model (postnatal 6 weeks)
(A) The number of NeuN+ cells decreased significantly in Arhgef2−/− mice brain cortex compared with wild-type mice. Red: NeuN. Blue: DAPI. WM: white matter. Scale bar, 250 μm, n = 6, ∗∗∗p < 0.001, unpaired t test.
(B) Representative images of Golgi staining. In Arhgef2−/− mice, morphology of neurites was abnormal. Scale bar, 250 μm.
(C) Total spines and mushroom spines significantly reduced in Arhgef2−/− mice compared with wild-type mice. The red arrow indicates mushroom spines. Scale bar, 5 μm. n = 3 per genotype, ∗∗p < 0.01, ∗p < 0.05, n.s.: no significant difference, unpaired t test.
(D) Compared with wild-type mice, expression of Psd-95 decreased significantly in Arhgef2−/− mice. n = 4 per genotype, ∗p < 0.05, unpaired t test. Data are represented as mean ± S.D.
Genetic deletion of Arhgef2 reduced m6A level in brain cortex
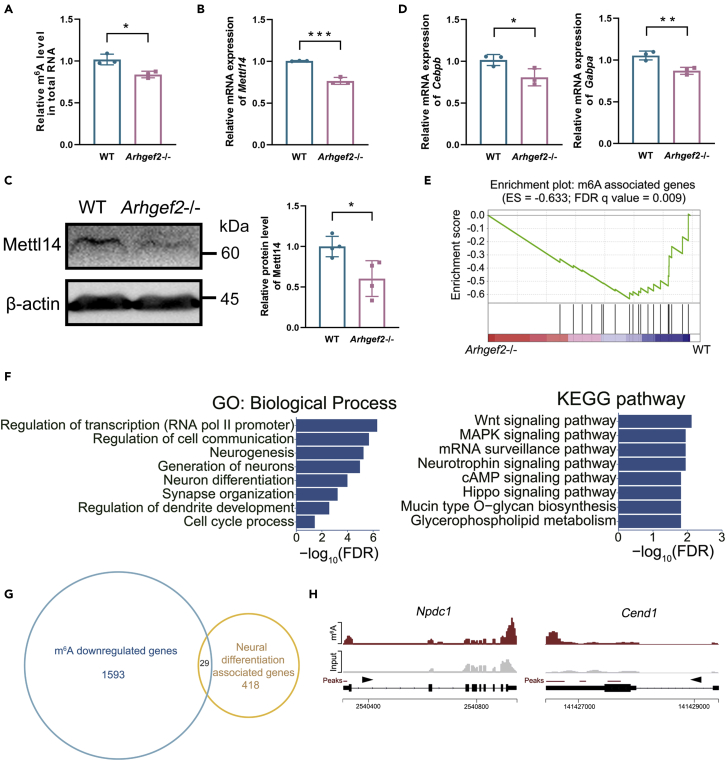
The present studies provided strong evidence that m6A deficiency in brain disrupted genes related to neuronal differentiation and impaired learning behaviors (Edens et al., 2019; Merkurjev, 2018; Koranda et al., 2018). To explore whether deficiency of Arhgef2 affects m6A level, we measured total m6A level of adult mouse brain cortex and the expression of writer and eraser genes involved in m6A methylation and demethylation. In Arhgef2−/− mice, total m6A level significantly decreased compared with wild-type mice. Meanwhile, mRNAs of methylases Mettl3 and Wtap showed a slight reduction without significance, whereas Mettl14, a vital factor controlling neural differentiation (Yoon et al., 2017), showed an observable downregulation. Demethylases Fto and Alkbh5 showed no observable change (Figures 3A–3C and S2A). Furthermore, we found Cebpb and Gabpa, transcription factors binding at the transcription start site of Mettl14, were significantly decreased in Arhgef2−/− mice compared with wild-type mice, whereas Cebpa, Elf1, and Spi1 showed no significant change (Weng et al., 2018) (Figures 3D and S2B). In parallel, Gene Set Enrichment Analysis (GSEA) of bulk RNA sequencing showed that the m6A-associated genes (writers, erasers, readers, Cebpb, and Gabpa) were significantly enriched in the downregulated genes of Arhgef2−/− mice (Figure 3E).
Figure 3.
Loss of Arhgef2 reduced m6A level and led to the enrichment of m6A-tagged mRNAs related to neural differentiation
(A) Total m6A level of mouse brain cortex was determined by m6A RNA Methylation Quantification Kit. n = 6, ∗p < 0.05, unpaired t test.
(B) The mRNA level of Mettl14 examined by qRT-PCR was significantly decreased in Arhgef2−/− mice compared with wild-type mice. n = 3 per genotype, ∗∗∗p < 0.001, unpaired t test.
(C) Representative western blot on brain cortex. Compared with wild-type mice, the protein expression of Mettl14 reduced in Arhgef2−/− mice. n = 4 per genotype, ∗p < 0.05.
(D) mRNAs of Cebpb and Gabpa, the translation factors of Mettl14, reduced significantly in Arhgef2−/− mice compared with wild-type mice. n = 3 per genotype (3 measurements for each sample), ∗p < 0.05, ∗∗p < 0.01, unpaired t test. Data are represented as mean ± SD.
(E) GSEA showed that the 21 m6A-associated genes (Mettl3, Mettl14, Mettl16, Wtap, Rbm15/15b, Kiaa1429, Fto, Alkbh5, Ythdc1/2, Ythdf1/2/3, eIF3, Abcf1, Fmr1, Igf2bp1/2/3, Cebpb, Gabpa) were significantly enriched in the downregulated genes of Arhgef2−/− mice. n = 3 per genotype, q < 0.05, GSEA software v3.1.0.
(F) GO analysis of 1,622 downregulated m6A-tagged genes revealed the enrichment for biological process terms related to transcription factors, cell communication, neurogenesis, synapse organization, and cell cycle. FDR, false discovery rate. Enrichment of KEGG pathway genes was also demonstrated.
(G) Venn diagram showing the overlap of m6A downregulated mRNAs (p < 0.05) and neural differentiation-associated genes.
(H) Coverage plot of m6A modification of Npdc1 and Cend1 mRNAs.
Loss of Arhgef2 inhibited the translation or nuclear export of m6A-tagged Npdc1 and Cend1 mRNAs related to neural differentiation
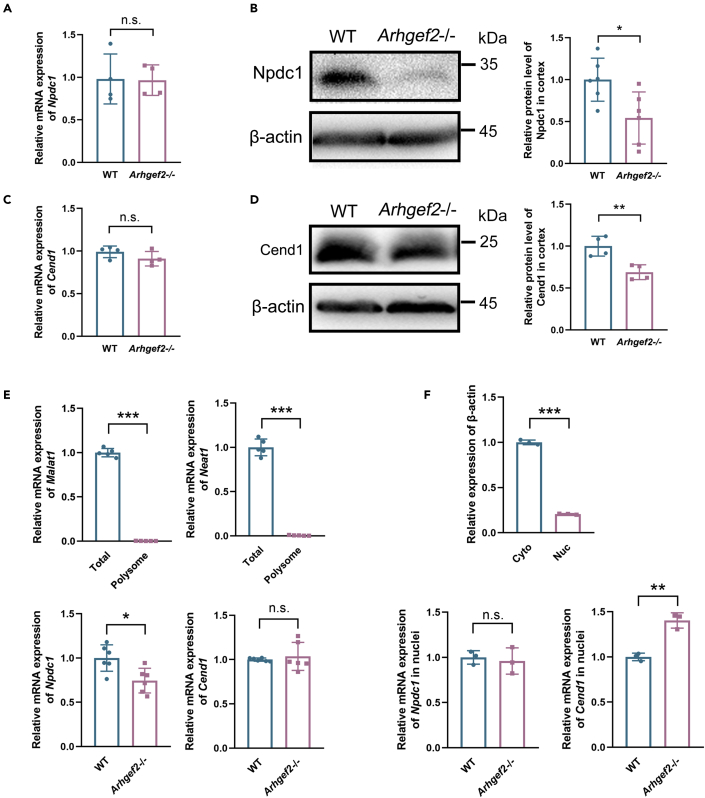
To explore how the deficiency of Arhgef2 affects neural differentiation by m6A modification, we performed m6A immunoprecipitation followed by RNA sequencing (m6A-seq). Similar patterns of the total m6A distribution in wild-type and Arhgef2−/− mice were observed, showing that m6A peaks were abundant in the vicinity of start and stop codons (Figure S3A). Loss of Arhgef2 resulted in the downregulation of 1,622 m6A-containing mRNAs and upregulation of 1,073 m6A-containing mRNAs. The cumulative distribution fraction of log2 (fold enrichment) demonstrated that m6A methylation of mRNAs tended to be lower in Arhgef2−/− mice than that in wild-type mice (Figure S3B). Gene Ontology (GO) and KEGG pathway analysis of the 1,622 m6A-tagged downregulated transcripts illustrated enrichment of genes related to cell communication, neurogenesis, neural differentiation, synapse organization, and cell cycle (Figures 3F and S3C). Interestingly, among 29 mRNAs that were both m6A significantly downregulated and neural differentiation-related, we found two of the mRNAs (Npdc1 and Cend1) strongly associated with cell cycle exit and terminal neural differentiation (Sansal et al., 2000; Politis et al., 2007) (Figures 3G and 3H). As Arhgef2 downregulation keeps radial precursors cycling, and thereby inhibits neurogenesis (Ravindran et al., 2017), we thus speculated that Npdc1 and Cend1 played a vital role in this process. Compared with wild-type mice, protein level of Npdc1 and Cend1 apparently decreased in Arhgef2−/− mice, whereas mRNAs showed no significant change by qRT-PCR (Figures 4A–4D), suggesting the positive effect of m6A modification on translation, nuclear export, or stability of both Npdc1 and Cend1. Interestingly, in Arhgef2−/− mice, whereas polysome profiling demonstrated the significant decrease of Npdc1 mRNA rather than Cend1 mRNA (Figure 4E), nuclear RNA extraction showed an increased expression of Cend1 mRNA, but not Npdc1 mRNA (Figure 4F). These results indicated that Arhgef2 mediated the translation of Npdc1 mRNA and nuclear export of Cend1 mRNA.
Figure 4.
Loss of Arhgef2 inhibited the translation of m6A-modified Npdc1 mRNA and nuclear export of m6A-modified Cend1 mRNA
(A–D) mRNA expression of Npdc1 and Cend1 showed no significant change determined by qRT-PCR. Protein levels of Npdc1 and Cend1 apparently decreased in Arhgef2−/− mice compared with wild-type ones. n.s.: no significant difference. n = 4–6 per genotype, ∗p < 0.05, ∗∗p < 0.01, unpaired t test.
(E) Npdc1 mRNA expression in polysome fractions significantly reduced in Arhgef2−/− mice compared with wild-type ones, but Cend1 mRNA showed no change. Poor expression of lncRNA (Malat1 and Neat1) in polysome fraction indicated the successful extraction of polysome fraction from other fractions. n.s.: no significant difference. ∗p < 0.05, ∗∗∗p < 0.001, unpaired t test. Total: total RNA from brain tissue. Polysome: polysome fraction RNA.
(F) Compared with wild-type mice, the nuclear Cend1 level, but not Npdc1, was significantly elevated in Arhgef2−/− mice. U1 was used as control gene in nuclei. Only low β-actin expression (~5-fold cytoplasm increase over nuclear) represented the successful separation of nuclear fractions. ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t test. n.s.: no significant difference. Cyto: cytoplasm. Nuc: Nuclei. Data are represented as mean ± S.D.
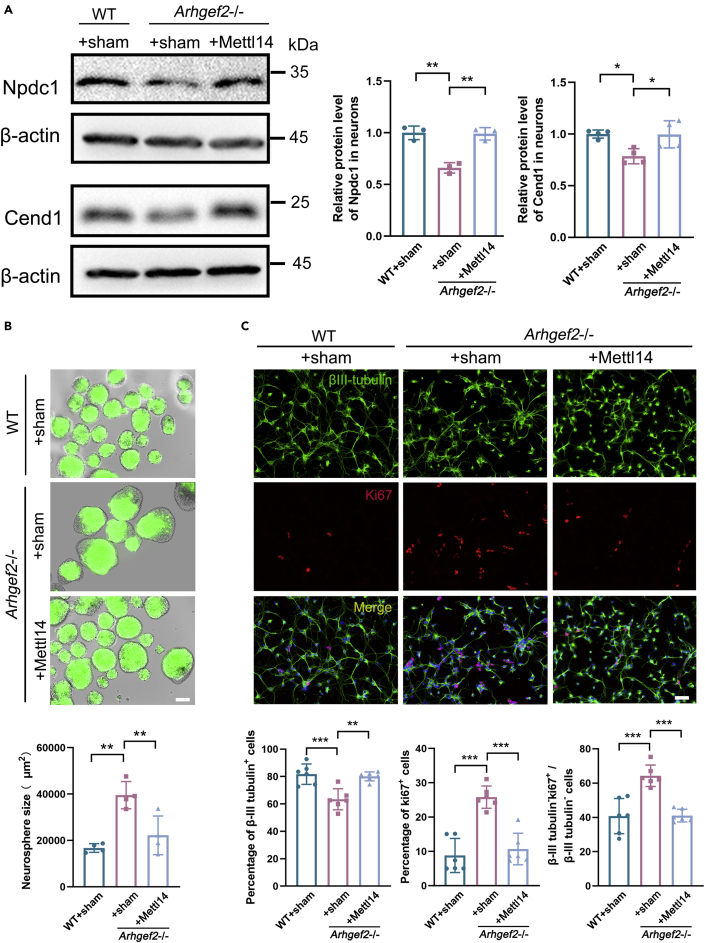
Overexpression of Mettl14 rescued neurogenesis and morphology of neurons during neural differentiation
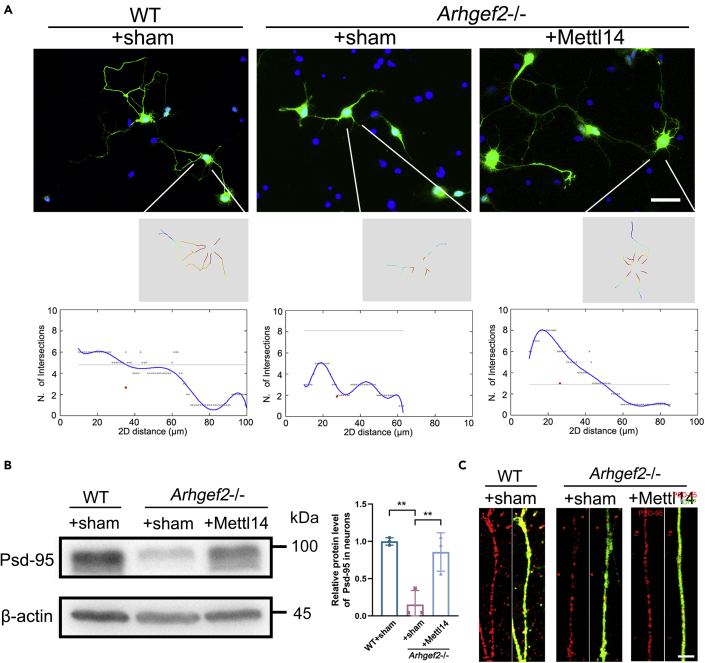
To characterize the impact of m6A on neural differentiation, primary cells of Arhgef2−/− mice were transfected with HBLV-m-Mettl14/Zsgreen (Figure S6). The total m6A level of primary neurons was significantly increased with overexpression of Mettl14 (Figure S4A). RNA immunoprecipitation (RIP) demonstrated that the enrichment of m6A-tagged Npdc1 and Cend1 mRNAs was significantly decreased in Arhgef2−/− RIP samples compared with wild-type RIP samples, which could be rescued after transfection with HBLV-m-Mettl14/Zsgreen (Figure S4B). Meanwhile, the distinct reduction of Npdc1 and Cend1 protein level in primary neurons from Arhgef2−/− mice also could be rescued by Mettl14 overexpression (Figure 5A). To reveal whether NPCs remain in cell cycle and fail to enter neural differentiation pathway, we measured the size of cortical neurospheres at 72 h after transfection. A significantly increased size of spheres was examined in Arhgef2−/− mice compared with wild-type mice, and the phenotypic anomaly could be recovered by HBLV-m-Mettl14/Zsgreen (Figure 5B). In addition, we identified a fewer number of βIII-tubulin+ cells (early neuron) and a larger proportion of βIII-tubulin-Ki67+ proliferating cells (precursors) in Arhgef2−/− mice than wild-type ones. Overexpression of Mettl14 was able to rescue this phenotype (Figure 5C). There was no significant variation in the proportion of phospho-histone H3 (pH3)-positive cells within 6 h of 5-ethynyl-2’-deoxyuridine (EdU) exposure (pH3+EdU+), suggesting that there was no defect in cell cycle to M phase by the loss of Arhgef2 (Figure S5). We also found that overexpression of Mettl14 rescued impaired neurite outgrowth and post-synaptic formation (Psd-95 as marker) in Arhgef2−/− mice (Figures 6A-6C).
Figure 5.
The anomaly of neurogenesis was rescued by overexpression of Mettl14 in primary NPCs from Arhgef2−/− mice (at E13.5)
(A) The protein expression of Npdc1 and Cend1 was examined by western blot. n = 3–4 per genotype, ∗p < 0.05, ∗∗p < 0.01, one-way ANOVA.
(B) The size of cortical neuro-spheres was measured at 72 h after transfection. A 2.3-fold increase of sphere size was noted by loss of Arhgef2, which could be recovered by HBLV-m-Mettl14-Zsgreen. Green: Zsgreen. Scale bar, 100 μm, n = 4 per genotype (3 measurements for each group), ∗∗p < 0.01, one-way ANOVA.
(C) Knockout of Arhgef2 significantly decreased βIII-tubulin+ cells (early neuron) and increased βIII-tubulin-Ki67+ proliferating cells (precursors), and the proportion could be rescued after Mettl14 overexpression. Cells were seeded at a density of 5 × 104/cm2 and were observed at day 4 after transfection. βIII-tubulin, marker for early neuron, green; Ki67, marker for proliferating cells, red; DAPI, blue. Scale bar, 50 μm. n = 6 per genotype, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA. Data are represented as mean ± SD.
Figure 6.
Overexpression of Mettl14 rescued the anomaly of neurite outgrowth and post-synaptic formation in primary neurons from Arhgef2−/− mice (at E13.5)
(A) The length of neurites and the number of branches of Zsgreen-positive cells was presented by using ImageJ combined with NeuronJ and Sholl Analysis plugin. Arhgef2 deficiency reduced the longest length of neurites and branch number. Cells were seeded at a density of 2.5 × 104/cm2 and were observed at day 3 after transfection. Scale bar, 25 μm.
(B) After transfection with HBLV-m-Mettl14-Zsgreen for 14 days, the expression of Psd-95 in primary matured neurons was measured by western blot analysis (left). Cells were seeded at a density of 1 × 105/cm2 and were observed at day 14 after transfection. Data are presented as means ± SD from three independent experiments (right), n = 3 per genotype, ∗∗p < 0.01, one-way ANOVA. Data are represented as mean ± SD.
(C) Immunostaining of Psd-95 in primary neurons. Psd-95 (marker for post-synapse, red), Zsgreen: green. Scale bar, 2.5 μm.
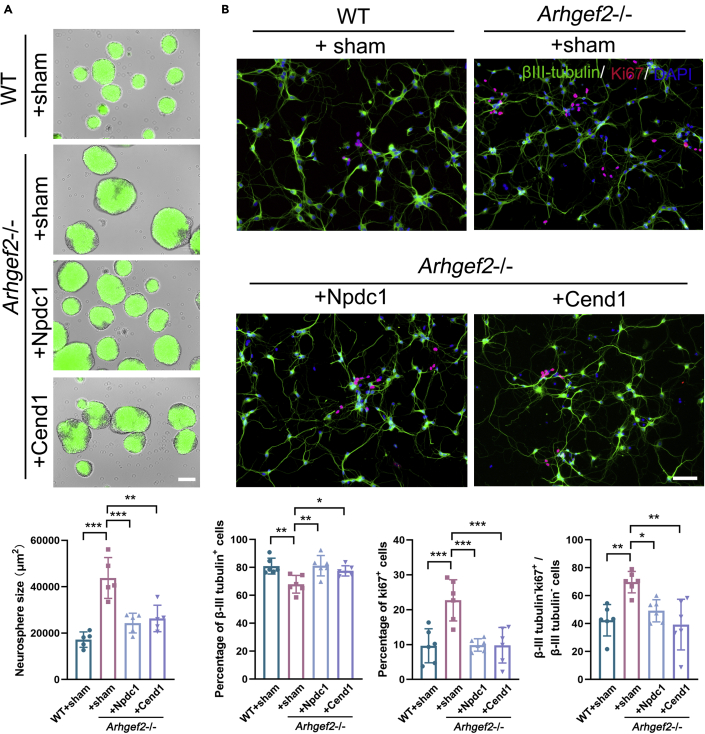
Overexpression of Npdc1 and Cend1 rescued neurogenesis and morphology of neurons during neural differentiation
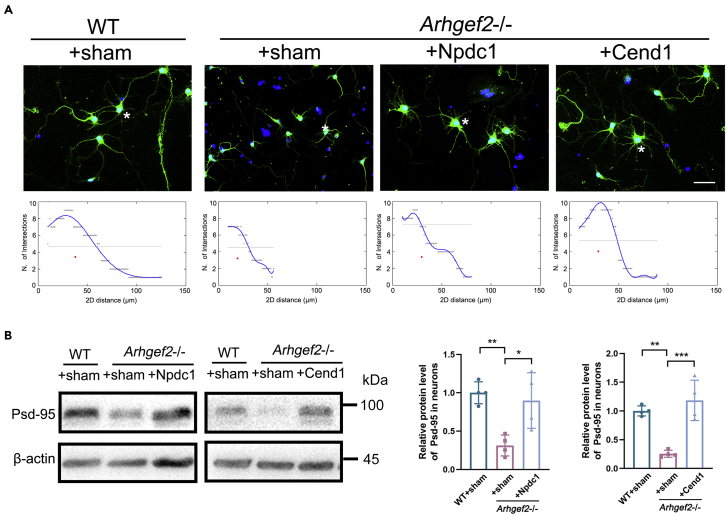
To further determine the role of Npdc1 and Cend1 in this process, we designed to overexpress Npdc1 in Arhgef2−/− mice (Figure S6). The measurement of neuro-spheres size suggested that Npdc1 promoted cell cycle exit (Figure 7A). An increased number of βIII-tubulin+ neurons and decreased number of βIII-tubulin-Ki67+ neurons were demonstrated in Arhgef2−/− mice transfected with HBLV-m-Npdc1/Zsgreen and HBLV-m-Cend1/Zsgreen, respectively (Figure 7B). Meanwhile, the abnormal morphology of neurite and expression of Psd-95 could also be, to some extent, rescued by overexpression of Npdc1 and Cend1 (Figures 8A and 8B).
Figure 7.
Overexpression of Npdc1 and Cend1 rescued the anomaly of neurogenesis in primary NPCs from Arhgef2−/− mice (at E13.5)
(A) Images of neuro-spheres showing a significant decrease size of spheres in Arhgef2-knockout NPCs after transfection with HBLV-m-Npdc1/Zsgreen and HBLV-m-Cend1/Zsgreen, respectively. Green: Zsgreen. Scale bar, 100 μm, n = 5 per genotype (2 measurements for each group), ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA.
(B) Both Npdc1 and Cend1 promoted the generation of βIII-tubulin+ cells (early neuron) and decreased βIII-tubulin-Ki67+ proliferating cells (precursors). Cells were seeded at a density of 5 × 104/cm2 and were observed at day 4 after transfection. βIII-tubulin, marker for early neuron, green; Ki67, marker for proliferating cells, red; DAPI, blue. Scale bar, 50 μm. n = 6 per genotype, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA. Data are represented as mean ± SD.
Figure 8.
Overexpression of Npdc1 and Cend1 rescued the anomaly of neurite outgrowth in primary neurons from Arhgef2−/− mice (at E13.5)
(A) Measurement of the length of neurites and the number of branches of Zsgreen-positive cells. Cells were seeded at a density of 2.5 × 104/cm2 and were observed at day 3 after transfection. Scale bar, 25 μm.
(B) After transfection with HBLV-m-Npdc1/Zsgreen and HBLV-m-Cend1/Zsgreen for 14 days, respectively, the expression of Psd-95 in primary matured neurons was measured by western blot analysis. Cells were seeded at a density of 1 × 105/cm2 and were observed at day 14 after transfection. n = 4 per genotype, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA. Data are represented as mean ± SD.
Discussion
m6A is emerging as a vital factor regulating neural differentiation (Yoon et al., 2017; Meyer et al., 2012). Here, we illustrated that loss of Arhgef2 reduced m6A level of brain cortical neurons. Specifically, methylases Mettl14 showed the significant reduction, suggesting its critical role in neural differentiation regulated by Arhgef2. Interestingly, defective Arhgef2 decreased m6A modification of target mRNAs, and thereby regulated not only neurogenesis but also neurite outgrowth and synaptic formation during neural differentiation. The abnormal phenotypes could be rescued by Mettl14.
As Arhgef2 down-regulation inhibits neurogenesis by keeping radial precursors cycling, and similar cellular phenotype is examined in Mettl14 knockout mice (Ravindran et al., 2017; Yoon et al., 2017), it strongly led us to focus on two m6A-modified target mRNAs (Npdc1 and Cend1), which were associated with cell cycle exit and terminal neural differentiation. Several studies indicate that Npdc1 is a neuronal differentiation regulator, which interacts with E2F-1 and inhibits cell cycle progression by affecting the functions of E2F-1 (Sansal et al., 2000; Evrard et al., 2004). Npdc1 positively regulates neurite outgrowth and involves in NRF-1-regulated neurite outgrowth (Tong et al., 2013). Cend1 (cell cycle exit and neuronal differentiation protein 1) coordinates cell cycle exit and differentiation of neuronal precursors (Politis et al., 2007). Deficiency of Cend1 impairs differentiation of certain types of neurons, including Purkinje cells (Sergaki et al., 2010). Both Npdc1 and Cend1 are expressed in dividing NPCs and are increased when cells start to differentiate. Their expression is developmentally regulated and persists in adults along the neuronal lineage (Dupont et al., 1998; Gaitanou et al., 2019). Arhgef2 deficiency decreased m6A methylations of the Npdc1 mRNA 5′ UTR and Cend1 mRNA 3′ UTR, which thus inhibited the translation of Npdc1 and nuclear export of Cend1 mRNAs. As overexpression of Mettl14 in primary neurons from Arhgef2−/− mice rescued m6A methylations of both target mRNAs, and thereby the protein level of Npdc1 and Cend1 was recovered near to wild-type ones, it suggested a significant impact of Mettl14-regulated m6A methylations on the translation of target mRNAs. Overexpression of Npdc1 and Cend1 rescued the anomaly of neurogenesis, neurite outgrowth, and synaptic formation, indicating their persistent critical role during neuronal differentiation.
Synaptic formation is regarded as a vital process in postnatal brain, which is involved in precise communication between neurons and other cellular partners. Psd-95, the excitatory post-synaptic marker, has been implicated in synaptic development and plasticity, and its defects disturb learning and memory (Yoo et al., 2019; Zhang and Lisman, 2012; Zheng et al., 2011; Grant, 2012). In our work, loss of Arhgef2 led to deficits in spine density and morphology and reduced expression of Psd-95 significantly. m6A sequencing of mouse cortex also revealed enrichment of mRNAs related to cell communication and synapse organization. Deficits of synaptic formation thus were a pivotal factor to explain the intellectual disability caused by Arhgef2 deficiency.
Arhgef2 catalyzes the replacement of GDP to GTP on RhoA (Birkenfeld et al., 2007). The GTP-bound form of RhoA causes Rho-associated kinase (ROCK) activation. Accumulating evidence point out that the RhoA/ROCK signaling participates in various neuronal functions, such as migration, dendrite development, and axonal extension (Fujita, 2014; Rico, 2004). In the last study, we reported that ARHGEF2-RhoA/ROCK pathway is essential for the migration and specification of precerebellar neurons. Specifically, the RhoA/ROCK/MLC pathway is most likely impaired in humans with the ARHGEF2 mutation (Ravindran et al., 2017). Several studies have indicated the role of RhoA/ROCK related to neural differentiation. NPCs with RhoA deficiency exhibit accelerated proliferation and reduction of cell cycle exit (Katayama et al., 2011). Activation of the RhoA/ROCK/JNK pathway could promote neuronal differentiation (Park et al., 2018).
However, whether ARHGEF2-RhoA/Rock pathway associated with m6A modification during neural differentiation is still unclear. We thus examined the expression of Mettl14 in wild-type primary neurons with treatment of Rock inhibitor Y27632. Surprisingly, mRNA and protein level of Mettl14 showed no significant change after treatment with Rock inhibitor, indicating that RhoA/Rock pathway maybe not implicated in Arhgef2/Mettl14-induced neuronal differentiation (Figure S7). Therefore, the exact pathway by which Arhgef2 regulates Mettl14 is worthy to be explored in the future work. Previous studies reported that Cebpb, Gabpa, Elf1, Cebpa, and Spi1 have binding sites within 5 kb upstream of the transcription start site of Mettl14. Among them, Gabpa and Elf1 showed a significant positive correlation, whereas Spil showed a significant negative correlation, with Mettl14 (Weng et al., 2018). In Arhgef2−/− mice, Cebpb and Gabpa were significantly reduced compared with wild-type mice, suggesting Arhgef2 was the most likely regulated expression of Mettl14 by them. This work elucidated a mechanism by which Arhgef2 deficiency inhibited the translation of m6A-modified Npdc1 and nuclear export of Cend1 mRNAs to impair neural differentiation. Intriguingly, it provided a considerable insight to think about the biological effects of the GEFs family on brain development associated with m6A modification.
Limitations of the study
This study linked Arhgef2 and neurogenesis via m6A methylation of target mRNAs, including those of Npdc1/Cend1, which are involved in cell cycle exit and neuronal differentiation. However, according to our present study and others' literatures, it is still hard to know how Arhgef2 affects Mettl14 through Cebpb and Gabpa. Arhgef2, a member of the Rho guanine nucleotide exchange factor family, is crucial for controlling the Ras superfamily. Cebpb is an essential downstream mediator of Ras signaling, which could be phosphorylated by Ras expression. We thus speculate that Arhgef2 may regulate the expression of Cebpb via Ras signaling. As for Gabpa, it is hard for us to speculate based on literatures.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-m6A | Millipore | Cat no: MABE1006 |
| Mouse monoclonal anti-NeuN | Millipore | Cat no: MAB377B; RRID: AB_177621 |
| Mouse monoclonal anti-β-III tubulin | Millipore | Cat no: MAB1637; RRID: AB_2210524 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat no: 9129; RRID: AB_2687446 |
| Rabbit polyclonal anti-Psd-95 | Cell Signaling Technology | Cat no: 3450;RRID: AB_2292883 |
| Mouse monoclonal anti-Synaptophysin | Abcam | Cat no: ab8049; RRID: AB_2198854 |
| Rabbit polyclonal anti-Mettl14 | Cell Signaling Technology | Cat no:51104S; RRID: AB_2799383 |
| Rabbit polyclonal anti-Npdc1 | Affinity | Cat no: DF4225; RRID: AB_2836576 |
| Rabbit monoclonal anti-Cend1 | Abcam | Cat no: ab113076; RRID: AB_10860676 |
| Rabbit polyclonal anti-Phospho-Histone H3 (Ser10) | Cell Signaling Technology | Cat no:9701; RRID: AB_331535 |
| Rabbit monoclonal anti-β-actin(D6A8) | Cell Signaling Technology | Cat no: 8457; RRID: AB_10950489 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat no: A-11036; RRID: AB_10563566 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat no: A-21236; RRID: AB_2535805 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat no: 7074; RRID: AB_2099233 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat no: 7076; RRID: AB_330924 |
| Deposited data | ||
| m6A-sequencing data | This paper | PRJNA730635 |
| Critical commercial assays | ||
| FD Rapid GolgiStainTM kit | Neuro Technologies | Cat no: PK401 |
| m6A RNA Methylation Quantification Kit (Colorimetric) | Abcam | Cat no: ab185912 |
| Imprint RNA Immunoprecipitation (RIP) Kit | Sigma-Aldrich | Cat no: RIP-12RXN |
| Click-iT EdU Imaging Kits | Thermo Fisher Scientific | Cat no: C10339 |
| Experimental models: Organisms/strains | ||
| Arhgef2 knockout mice | Shanghai Biomodel Organism Science & Technology Development | N/A |
| Oligonucleotides | ||
| β-actin: (F) 5’-GGCTGTATTCCCCTCCATCG-3’ (R) 5’-CCAGTTGGTAACAATGCCATGT-3’ |
This paper | N/A |
| Arhgef2: (F) 5’-ATCTCCGTCTCCGGCATGA-3’ (R) 5’-CAGGGTGTCTTTACAGCGGTT-3’ |
This paper | N/A |
| Npdc1: (F) 5’-CTTCAACACCACGAATCTCGC-3’ (R) 5’-CCTTTGGAGGTTCCTTATGCC-3’ |
This paper | N/A |
| Cend1: (F) 5’-ACCAGCCAAGGCAGATCCT-3’ (R) 5’-GGTCAAGTTCTCACAAGGCCA-3’ |
This paper | N/A |
| Mettl14: (F) 5’-TCTGAGAGTGCGGATAGCATTG-3’ (R) 5’-GAGCAGATGTATCATAGGAAGCC-3’ |
This paper | N/A |
| Mettl3: (F) 5’-CTGGGCACTTGGATTTAAGGAA-3’ (R) 5’-TGAGAGGTGGTGTAGCAACTT-3’ |
This paper | N/A |
| Wtap: (F) 5’-ATGGCACGGGATGAGTTAATTC-3’ (R) 5’-TTCCCTTAAACCAGTCACATCG-3’ |
This paper | N/A |
| Fto: (F) 5’-CCGTCCTGCGATGATGAAGT-3’ (R) 5’-CCCATGCCGAAATAGGGCTC-3’ |
This paper | N/A |
| Alkbh5: (F) 5’-TTGCCACCCAGCTATGCTTC-3’ (R) 5’-CAGACCGCCGGTTTTCTTCTT-3’ |
This paper | N/A |
| Gabpa: (F) 5’-AGCGCATCTCGTTGAAGAAG-3’ (R) 5’-TCCTGCTCTTTTCTGTAGCCT-3’ |
This paper | N/A |
| Cebpb: (F) 5’-CAAGATGCGCAACCTGGAGA-3’ (R) 5’-GACAGCTGCTCCACCTTCTT-3’ |
This paper | N/A |
| Cebpa: (F) 5’-CAAGAACAGCAACGAGTACCG-3’ (R) 5’-GTCACTGGTCAACTCCAGCAC-3’ |
This paper | N/A |
| Elf1: (F) 5’-TGTCCAACAGAACGACCTAGT-3’ (R) 5’-CACACAAGCTAGACCAGCATAA-3’ |
This paper | N/A |
| Spi1: (F) 5’-ATGTTACAGGCGTGCAAAATGG-3’ (R) 5’-TGATCGCTATGGCTTTCTCCA-3’ |
This paper | N/A |
| Malat1: (F)5’-CATGGCGGAATTGCTGGTA-3’ (R)5’-CGTGCCAACAGCATAGCAGTA-3’ |
This paper | N/A |
| Neat1: (F)5’-TGGCCCCTTTTGTTCATTAGC-3’ (R)5’-TGGAAGGCCATTGTTTCAGG-3’ |
This paper | N/A |
| U1: (F) 5’-GATACCATGATCACGAAGGTGGTT-3’ (R) 5’-CACAAATTATGCAGTCGAGTTTCC-3’ |
This paper | N/A |
| Recombinant DNA | ||
| HBLV-m-mettl14-3xflag-Zsgreen-PURO | Hanbio | N/A |
| HBLV-m-Npdc1-3xflag-Zsgreen-PURO | Hanbio | N/A |
| HBLV-m-Cend1-3xflag-Zsgreen-PURO | Hanbio | N/A |
| Software and algorithms | ||
| Prism 7 | GraphPad | SCR_002798; https://www.graphpad.com |
| ImageJ | Wayne Rasband National Institutes of Health | https://imagej.nih.gov/ij/ |
| Fastp | Chen et al., 2018 | https://github.com/OpenGene/fastp |
| IGV | Meng et al., 2014 | http://www.igv.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hao Hu (huh@cougarlab.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Sequencing data have been deposited with the Sequence Read Archive (SRA) under BioProject :PRJNA730635(https://www.ncbi.nlm.nih.gov/bioproject/PRJNA730635).
Experimental model and subject details
Mice model establishment
All mice experiments in this work were approved by the institutional animal care and use committee in the Guangzhou Medical University (registration No. 2019-436, 2019-694). Arhgef2 knockout mice were created using CRISPR/Cas9-mediated genome engineering. Exon 2 was selected as the target site, and gRNA was designed as following sequences: gRNA1: 5’-ACCTACCTGACCCCGGCGACTGG-3’; gRNA2: 5’-TATTGGAATGTGGCGTAAGCTGG-3’.
The zygotes co-injected with Cas9 mRNA and gRNA were cultured in medium until the two-cell stage, and then were transferred into the oviducts of pseudopregnant ICR females. After being identified by genomic PCR and Sanger sequencing, the mutant F0 were bred with C57BL/6J males to yield heterozygous Arhgef2 mice. Subsequent breeding of the heterozygous Arhgef2 mice generated offspring with wild-type and knockout littermates for experiments. For animal experiments, Arhgef2 knockout male and female mice (sex balance) at age of 6-8 weeks were used. For primary cell culture, Arhgef2 heterozygous pregnant mice were used.
Cell culture
Primary mouse NPCs were isolated from Arhgef2 wild-type and knockout mice embryonic cortices at E13.5 and cultured in Neurobasal medium (Gibco, CA, USA) containing 2% B27 (v/v, Gibco), 0.25% Glutamax, and 25 μM glutamate. Cells were seeded on dishes precoated with poly-D-lysine. To measure neuro-spheres, NPCs were cultured in ultra-low attachment plates and the size pf spheres was analyzed by Image J. The experiments of neurogenesis were performed on density-matched NPCs cultures within the first 4 days, while the experiments related to synaptic formation were conducted in well-matured neurons at day 14. For the overexpression of Mettl14 or Npdc1, transfection of HBLV-m-Mettl14/Zsgreen and HBLV-m-Npdc1/Zsgreen (at MOI 10) (Hanbio Biotechnology, Shanghai, China) respectively to Arhgef2-knockout primary cells was performed. HBLV-Zsgreen was transfected to wild-type and Arhgef2 -knockout cells as control.
Method details
Tissue preparation
Mice at 6 weeks of age were anaesthetized with 50 mg/kg sodium pentobarbital. After perfused with 0.9% NaCl and 4% PFA, brains were harvested and fixed in 4% PFA overnight, followed with dehydration by 15% and 30% sucrose. Sagittal brain sections (30 μm thick) were prepared using freezing microtome (CM3050S, Leica, Germany).
Nissl staining
Brain sections were treated with 75% ethanol for 30 s, dH2O for 30 s and cresyl violet for 2 min. After dehydration with gradient ethanol (75%, 95%, 100%), sections were incubated with xylene and mounted using neutral resins. The images were taken under microscope.
Immunostaining
To determine the number of matured neurons in brain cortex, sections were permeabilized with 0.3% Triton X-100 and blocked with goat serum for 1 h at room temperature, followed by incubation with mouse anti-NeuN antibody at 4°C overnight. After washing, HRP-conjugated anti-mouse IgG secondary antibody was applied to the sections for 1 h at room temperature. Nuclei were stained using 4,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific).
To examine the neurogenesis of NPCs, cells were immersed in 4% PFA for fixation, 0.3% Triton X-100 for permeabilization, and then blocking. Cells were incubated with mouse anti-β-III tubulin antibody and rabbit anti-ki67 antibody overnight, followed by secondary antibodies. To observe the synaptic formation, Zsgreen-positive cells were co-staining with Psd-95 as above. Immunostaining of pH3 in primary cells exposed to EdU was also performed as above. Data were obtained from at least three independent experiments and analyzed by ImageJ software.
Golgi staining
Brain tissue prepared for Golgi staining was performed by using FD Rapid GolgiStainTM kit. According to manufacturer’s instruction, tissue was immersed in Solution A/B at room temperature for at least 24 h, followed by Solution C. All procedures should be protected from light. Samples embedded in OCT compound were then sectioned coronally (30 μm thick). For staining procedure, sections were placed in Solution D/E for 10 min. After washing, sections were dehydrated in 50%, 75%, and 95% ethanol, followed by 100% ethanol 4 times for 4 min each rinse. Sections then were cleared by xylene and mounted. The confocal imaging was achieved using a TCS-SP8 LSM confocal imaging system (Leica, Wetzlar, Germany).
Quantification of the m6A
The m6A level of brain cortex was measured using m6A RNA Methylation Quantification Kit (Colorimetric). In brief, total RNA of brain cortex was isolated using TRIZOL reagent (Invitrogen, Thermo Fisher Scientific, USA). 100 -300 ng total RNA per reaction was added into the wells filled with 80 μl Binding Solution. After incubation at 37°C for 90 min, wells were washed with 1X Wash Buffer. m6A RNA was then captured using Diluted Capture Antibody, following by Diluted Detection Antibody and Diluted Enhancer Solution. Developer Solution was added to each well and incubated for 1-10 min away from light. Signal was detected on a microplate reader at 450 nm.
Quantitative real-time PCR (qPCR)
Total RNA was extracted from the mice brain tissues using TRIZOL reagent (Invitrogen, Thermo Fisher Scientific, MA, USA), and first-strand cDNA was synthesized with PrimeScript RT reagent (Takara-Clonetech, Shiga, Japan). qRT-PCR was performed using SYBR Premix Ex TaqII (Takara-Clonetech) and analyzed with Fast Real Time System 7500 (Applied Biosystems). Expression levels of each target gene were measured relative to β-actin as internal control. The following sequences of primers were exhibited in Table (Table S1). The experiments were repeated at least three times, each time with consistent results.
m6A-sequencing
Total RNA was isolated from the cortex samples of the WT and KO mice and purified using TRIzol reagent following the manufacturer's procedure. The RNA integrity was determined and confirmed using Bioanalyzer 2100 (Agilent, CA, USA) with RIN number >7.0 and electrophoresis with denaturing agarose gel, respectively. Using Dynabeads Oligo (dT) 25-61005 (Thermo Fisher, CA, USA), Poly (A) RNA was purified from 50 μg total RNA under two rounds of purification. Under 86°C, the poly (A) RNA was fragmented into small pieces using Magnesium RNA Fragmentation Module (NEB, cat. e6150, USA) for 7 min. m6A pull-down was performed using m6A-specific antibody (No. 202003, Synaptic Systems, Germany) in IP buffer (50 mM Tris-HCl, 750 mM NaCl and 0.5% Igepal CA-630) for 2 h at 4°C. Next, the IP RNA was reverse-transcribed to cDNA, followed by synthesis of U-labeled second-stranded DNAs with E. coli DNA polymerase I (NEB, cat.m0209, USA), RNase H (NEB, cat.m0297, USA) and dUTP Solution (Thermo Fisher, cat. R0133, USA).
An A-base was then added to the blunt ends of each strand, which was prepared for ligation to the indexed adapters. Single- or dual-index adapters were ligated to the fragments; size selection was performed with AMPureXP beads. After the heat-labile UDG enzyme (NEB, cat.m0280, USA) treatment, the ligated products were amplified with PCR. The average insert size for the final cDNA library was 300 ± 50 bp. The 2 ×150 bp paired-end sequencing (PE150) was performed on an illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China).
Bioinformatic analyses of m6A-seq
The reads containing adaptor contamination, low quality bases and undetermined bases were removed using the fastp software (Chen et al., 2018) with default parameter. Sequence quality of IP and Input samples was verified by Fastp. The reads were then mapped to the reference genome Mus musculus (Version: v99) using HISAT2 (Kim et al., 2015). The m6A peaks with bed or bigwig format that can be adapted for visualization on the IGV software (http://www.igv.org) were identified by the R package exomePeak (Meng et al., 2014). MEME (Bailey et al., 2009) and HOMER (Heinz et al., 2010) were used for de novo and known motif finding, followed by localization of the motif with respect to peak summit. Called peaks were annotated using R package ChIPseeker (Yu et al., 2015). The expression level for all mRNAs from input libraries was determined using StringTie (Pertea et al., 2015) by calculating FPKM (total exon fragments /mapped reads (millions) × exon length (kB)). Gene ontology analysis was performed on the 1622 m6A downregulated genes using GO enrichment analysis tool of Omicshare. KEGG pathway analysis was also performed on the 1622 m6A downregulated genes using pathway enrichment analysis tool of Omicshare. GO terms and KEGG pathways with FDR corrected p < 0.05 was considered significant. Protein-protein interaction network was obtained from the STRING database (Szklarczyk et al., 2017). The interaction network model was generated using Cytoscape 3.6.1 (Shannon et al., 2003). The lastest gene list was downloaded from the BioMart database (https://www.ensembl.org/biomart/martview/) where 447 neural differentiation associated genes were identified according to their functional annotation and related GO terms.
Neurite outgrowth analysis
Neurite outgrowth of Zsgreen-positive cells was measured using Image J combined with Neuron J and Sholl Analysis plugin. A vertical line from the soma to the tip of the longest branch was drawn to defined as the length of neurite. the number of branches was examined by counting the intersections of neurites and concentric circles. The radius interval between circles was 10 μm per step, ranging from 10 μm to the tips of the longest branch.
EdU staining
Primary cells were labeled with 10 μM EdU solution and incubated for 4 h at 37°C and then fixed with 4% PFA. After permeabilization with 0.5% TritonX-100, cells were treated with Click-iT EdU Imaging Kits according to manufacturer’s protocol.
RNA immunoprecipitation (RIP)
After transfection of lentivirus for 3 days, m6A modification on Npdc1 and Cend1 mRNA was detected using Imprint RNA Immunoprecipitation (RIP) Kit. In brief, cells (1-2 X 106 each reaction) were collected in mild lysis buffer supplemented with protease inhibitor cocktail, 1 M DTT and 40 U/μl ribonuclease inhibitor. After incubation on ice for 15 min, the lysis reactions were centrifuged for 10 min at 16,000 g at 4°C, and supernatant was collected. 10% of RIP lysate supernatant per reaction was labelled as Input. Each reaction was then incubated with 2 μg of anti-m6A antibody-prebinding protein A magnetic beads on a rotator overnight. 2 μg of IgG antibody was used for negative control. m6A-tagged mRNA was eluted by IP buffer and purified using TRIZOL reagent. SYBR-green based qRT-PCR was performed as described above.
Western blot
Protein was extracted using 2% SDS containing protease inhibitor cocktail and phosphatase inhibitor (Cell Signaling Technology, MA, USA). Samples were separated with 12.5% SDS-PAGE and transferred to PVDF membranes (Millipore, MA, USA). The membranes were blocked in 5% skim milk for 1 h at room temperature and then incubated overnight at 4°C with the following primary antibodies: rabbit anti-Npdc1 antibody, rabbit anti-Cend1 antibody, rabbit anti-Mettl14 antibody, rabbit anti-Psd-95 antibody, and mouse anti-Synaptophysin antibody, respectively. After washing with TBST, the membranes were incubated with secondary antibodies for 1 h at room temperature. Blots were visualized by ChemiDocTM MP system (Bio-Rad, USA) and then quantified via ImageJ software.
Polysome profiling
10%~45% sucrose density gradients were prepared using sucrose gradient buffer (10 mM HEPES-KOH, 5 mM MgCl2, 150 mM KCl, 200 U/ml RNase inhibitor, 1x protease inhibitor cocktail, 100 μg/ml cycloheximide). Brain tissue was grinded in a liquid nitrogen-proof container to obtain powder. The powder was collected in a modified lysis buffer (10 mM HEPES-KOH, 5 mM MgCl2, 150 mM KCl, 0.5 mM DTT, 100 U/ml RNase inhibitor, 1x protease inhibitor cocktail, 100 μg/ml cycloheximide, 0.5% NP-40, 0.05% Triton-X100) and further homogenized using electric grinding rod. The homogenate was centrifuged at 20000 rcf for 10 min at 4°C or reserved for total RNA isolation. The supernatant was layered onto sucrose gradient and centrifuged in a SW-40Ti rotor at 36,000 rpm for 2.5 h at 4°C and then analyzed using Gradient Fractionator (Biocomp). Polysome fraction was collected and RNA was extracted with TRIzol LS Reagent (Invitrogen, MA, USA) according to manufacturer’s instructions. qRT-PCR was performed as described above. LncRNA (Malat1 and Neat1) was used as quality control of polysome fraction.
Nuclear RNA extraction
Homogenization of mice brain tissue with liquid nitrogen grinding, and nuclear fractions were prepared using NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. 100U/ml of RNase inhibitor (Ambon/Fisher) was added to the CERI reagent to prevent RNA degradation. After separation from the cytoplasmic fraction, the nuclear fraction was washed twice with ice PBS (prepared in RNase-free water), and resuspended in an appropriate volume of TRIzol reagent (Invitrogen). RNA was purified using the TRIzol reagent manufacturer’s protocol. The efficiency of separation was determined by measuring nuclear and cytoplasmic RNA fractions by qRT-PCR for the ratio of β-actin expression in nuclei and cytoplasm. Only nuclear RNA preparations with low β-actin expression (~5-fold cytoplasm increase over nuclear) were used for analysis. To compare the expression of Npdc1 and Cend1 mRNA in nuclei, U1 was used as control gene.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism8 software. All graphs present mean ± S.D. Statistical comparisons of the data were carried out by student’s t-test and One-way ANOVA. P<0.05 was considered statistically significant and significant level is indicated as follows: ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001. Specific details including the statistic test and the number of experimental replicates were demonstrated in figure legends.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81671067, 81974163, and 82001121), the Major Medical Collaboration and Innovation Program of Guangzhou Science Technology and Innovation Commission (201604020020); the China Postdoctoral Science Foundation (2019M662852); and the Key-Area Research and Development Program of Guangdong Province (2019B020227001).
Author contributions
H.H. conceived and coordinated the project. P.Z. and N.L. oversaw the mice modeling with aids of S.Z., C.L., and L.L. P.Z. and Y.Q. designed and performed most of the experiments with aids of M.Y., S.Z., F.Q., H.L., Y.L., E.R., C.S., D.H., C.P., and N.L. X.F. was in charge of the genetic analysis and bioinformatics with aids of X.W. P.Z., and Y.Q. wrote the manuscript with aids of H.H., N.L., and A.M.K.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102645.
Contributor Information
Yifei Qi, Email: eddy3207@126.com.
Hao Hu, Email: huh@cougarlab.org.
Supplemental information
References
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal C.J., Finan D., LaRose J., Wells C.D., Gish G., Kulkarni S., DeSepulveda P., Wilde A., Rottapel R. The Rho GTP exchange factor Lfc promotes spindle assembly in early mitosis. Proc. Natl. Acad. Sci. USA. 2005;102:9529–9534. doi: 10.1073/pnas.0504190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenfeld J., Nalbant P., Bohl B.P., Pertz O., Hahn K.M., Bokoch G.M. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev. Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y.Q., Chen Y., Jia G. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont E., Sansal I., Evarard C., Rouget P. Developmental pattern of expression of NPDC-1 and its interaction with E2F-1 suggest a role in the control of proliferation and differentiation of neural cells. J. Neurosci. Res. 1998;51:257–267. doi: 10.1002/(SICI)1097-4547(19980115)51:2<257::AID-JNR14>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Edens B.M., Vissers C., Su J., Arumugam S., Xu Z., Shi H., Miller N., Rojas Ringeling F., Ming G.L., He C. FMRP modulates neural differentiation through m6A-dependent mRNA nuclear export. Cell Rep. 2019;28:845–854.e5. doi: 10.1016/j.celrep.2019.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard C., Caron S., Rouget P. Functional analysis of the NPDC-1 gene. Gene. 2004;343:153–163. doi: 10.1016/j.gene.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Yamashita T. Axon growth inhibition by RhoA/ROCK in the central nervous system. Front. Neurosci. 2014;8:338. doi: 10.3389/fnins.2014.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanou M., Segklia K., Matsas R. Cend1, a story with many tales: from regulation of cell cycle progression/exit of neural stem cells to brain structure and function. Stem Cells Int. 2019;2019:2054783. doi: 10.1155/2019/2054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.G. Synaptopathies: diseases of the synaptome. Curr. Opin. Neurobiol. 2012;22:522–529. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Haussmann I.U., Bodi Z., Sanchez-Moran E., Mongan N.P., Archer N., Fray R.G., Soller M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;540:301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama K., Melendez J., Baumann J.M., Leslie J.R., Chauhan B.K., Nemkul N., Lang R.A., Kuan C.Y., Zheng Y., Yoshida Y. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proc. Natl. Acad. Sci. USA. 2011;108:7607–7612. doi: 10.1073/pnas.1101347108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda J.L., Dore L., Shi H., Patel M.J., Vaasjo L.O., Rao M.N., Chen K., Lu Z., Yi Y., Chi W. Mettl14 is essential for Epitranscriptomic regulation of striatal function and learning. Neuron. 2018;99:283–292.e5. doi: 10.1016/j.neuron.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M., Zenke F.T., Bokoch G.M. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat. Cell. Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- Li Q., Li X., Tang H., Jiang B., Dou Y., Gorospe M., Wang W. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J. Cell. Biochem. 2017;118:2587–2598. doi: 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri D., Marshall C.B., Greeve M.A., Kim B., Balan M., Suarez F., Bakal C., Wu C., Larose J., Fine N. Mechanistic insight into the microtubule and actin cytoskeleton coupling through dynein-dependent RhoGEF inhibition. Mol. Cell. 2012;45:642–655. doi: 10.1002/jcb.25957. [DOI] [PubMed] [Google Scholar]
- Meng J., Lu Z., Liu H., Zhang L., Zhang S., Chen Y., Rao M.K., Huang Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkurjev D., Hong W.T., Iida K., Oomoto I., Goldie B.J., Yamaguti H., Ohara T., Kawaguchi S.Y., Hirano T., Martin K.C. Synaptic N6-methyladenosine (m6A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 2018;21:1004–1014. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Meyer K.D., Jaffrey S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 30 UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbant P., Chang Y.C., Birkenfeld J., Chang Z.F., Bokoch G.M. Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol. Biol. Cell. 2009;20:4070–4082. doi: 10.1091/mbc.e09-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Kang M.J., Han J.S. Interleukin-1 beta promotes neuronal differentiation through the Wnt5a/RhoA/JNK pathway in cortical neural precursor cells. Mol. Brain. 2018;11:39. doi: 10.1186/s13041-018-0383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis P.K., Makri G., Thomaidou D., Geissen M., Rohrer H., Matsas R. BM88/CEND1 coordinates cell cycle exit and differentiation of neuronal precursors. Proc. Natl. Acad. Sci. USA. 2007;104:17861–17866. doi: 10.1073/pnas.0610973104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran E., Hu H., Yuzwa S.A., Hernandez-Miranda L.R., Kraemer N., Ninnemann O., Musante L., Boltshauser E., Schindler D., Hübner A. Homozygous ARHGEF2 mutation causes intellectual disability and midbrain-hindbrain malformation. PLoS Genet. 2017;13:e1006746. doi: 10.1371/journal.pgen.1006746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Li R., Zheng Y., Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J. Biol. Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- Rico B., Beggs H.E., Schahin-Reed D., Kimes N., Schmidt A., Reichardt L.F. Control of axonal branching and synapse formation by focal adhesion kinase. Nat. Neurosci. 2004;7:059–69. doi: 10.1038/nn1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansal I., Dupont E., Toru D., Evrard C., Rouget P. NPDC-1, a regulator of neural cell proliferation and differentiation, interacts with E2F-1, reduces its binding to DNA and modulates its transcriptional activity. Oncogene. 2000;19:5000–5009. doi: 10.1038/sj.onc.1203843. [DOI] [PubMed] [Google Scholar]
- Sergaki M.C., Guillemot F., Matsas R. Impaired cerebellar development and deficits in motor coordination in mice lacking the neuronal protein BM88/Cend1. Mol. Cell Neurosci. 2010;44:15–29. doi: 10.1016/j.mcn.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonami K., Kurihara Y., Arima S., Nishiyama K., Uchijima Y., Asano T., Sorimachi H., Kurihara H. Calpain-6, a microtubule-stabilizing protein, regulates Rac1 activity and cell motility through interaction with GEF-H1. J. Cell. Sci. 2011;124:1214–1223. doi: 10.1242/jcs.072561. [DOI] [PubMed] [Google Scholar]
- Tong C.W., Wang J.L., Jiang M.S., Hsu C.H., Chang W.T., Huang A.M. Novel genes that mediate nuclear respiratory factor 1-regualted neurite outgrowth in neuroblastoma IMR-32 cells. Gene. 2013;515:62–70. doi: 10.1016/j.gene.2012.11.026. [DOI] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhao J.C. Update: mechanisms underlying N6-methyladenosine modification of eukaryotic mRNA. Trends Genet. 2016;32:763–773. doi: 10.1016/j.tig.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., Shi H., Skibbe J., Shen C., Hu C. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo K.S., Lee K., Oh J.Y., Lee H., Park H., Park Y.S., Kim H.K. Postsynaptic density protein 95 (PSD-95) is transported by KIF5 to dendritic regions. Mol. Brain. 2019;12:97. doi: 10.1186/s13041-019-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki H., Ohba Y., Parrini M.C., Dulyaninova N.G., Bresnick A.R., Mochizuki N., Matsuda M. Cell type-specific regulation of RhoA activity during cytokinesis. J. Biol. Chem. 2004;279:44756–44762. doi: 10.1074/jbc.M402292200. [DOI] [PubMed] [Google Scholar]
- Zhang P., Lisman J.E. Activity-dependent regulation of synaptic strength by PSD-95 in CA1 neurons. J. Neurophysiol. 2012;107:1058–1066. doi: 10.1152/jn.00526.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C.Y., Seabold G.K., Horak M., Petralia R.S. MAGUKs, synaptic development, and synaptic plasticity. Neuroscientist. 2011;17:493–512. doi: 10.1177/1073858410386384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y.Y., Liu H.J., Song X., Ju Y., Peng H. A linear regression predictor for identifying N(6)-Methyladenosine sites using frequent gapped K-mer pattern. Mol. Ther. Nucleic Acids. 2019;18:673–680. doi: 10.1016/j.omtn.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited with the Sequence Read Archive (SRA) under BioProject :PRJNA730635(https://www.ncbi.nlm.nih.gov/bioproject/PRJNA730635).