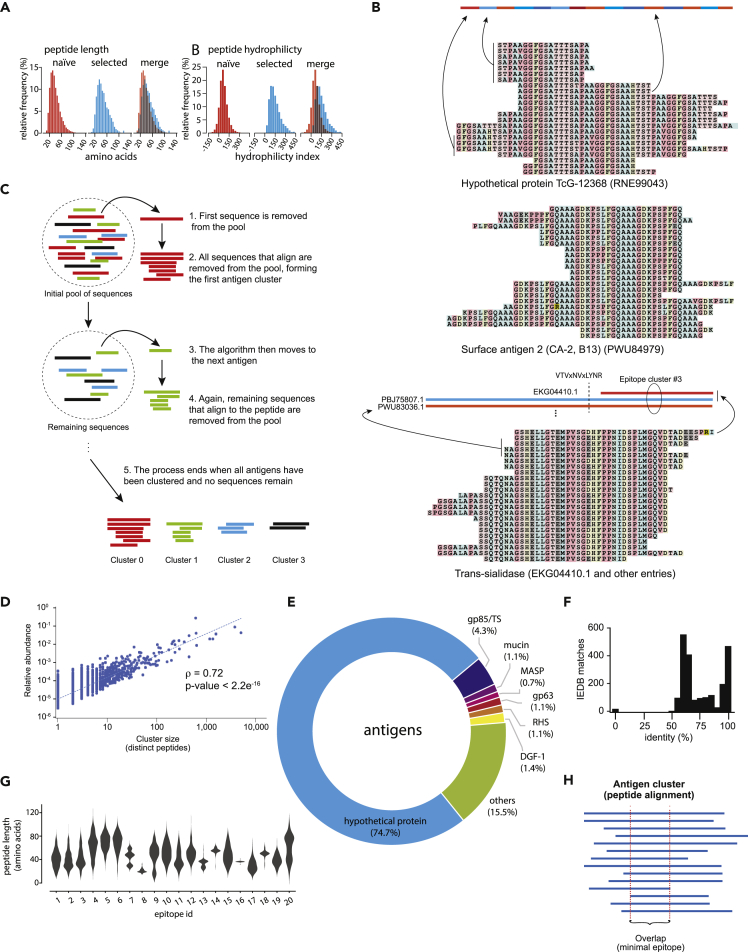
Figure 3.
Antigen identification and epitope mapping
(A) The plots show peptide distribution as per length (left) and hydrophilicity (right) in the naive library (red) and after three rounds of selection with IgG (blue) of patients with Chagas disease
(B) Alignment illustrating three clusters of epitopes. Each line represents a unique T. cruzi-derived peptide identified in the final round of selection. The top two clusters comprised epitopes belonging to repetitive sequences of the same protein. The bottom one is a cluster that maps to an epitope shared by distinct members of the gp85/trans-sialidase multigene family.
(C) Scheme to illustrate the algorithm used to cluster all unique sequences identified in the third round of the biopanning to identify the 3,964 epitope/antigens pairs.
(D) Scatchard plot showing the correlation between antigen cluster abundance x number of peptides in the cluster (Spearman correlation).
(E) Pie chart displaying all final antigens and their corresponding similarity to T. cruzi protein families.
(F) Bar graph indicating the number of antigen clusters that match previously described T. cruzi antigens (IEDB).
(G) Violin plot displaying the length and density of selected clusters of antigens.
(H) Cartoon illustrating our epitope mapping strategy. After clusterization, all sequences were aligned and the minimal overlapping region shared by all peptides was identified.