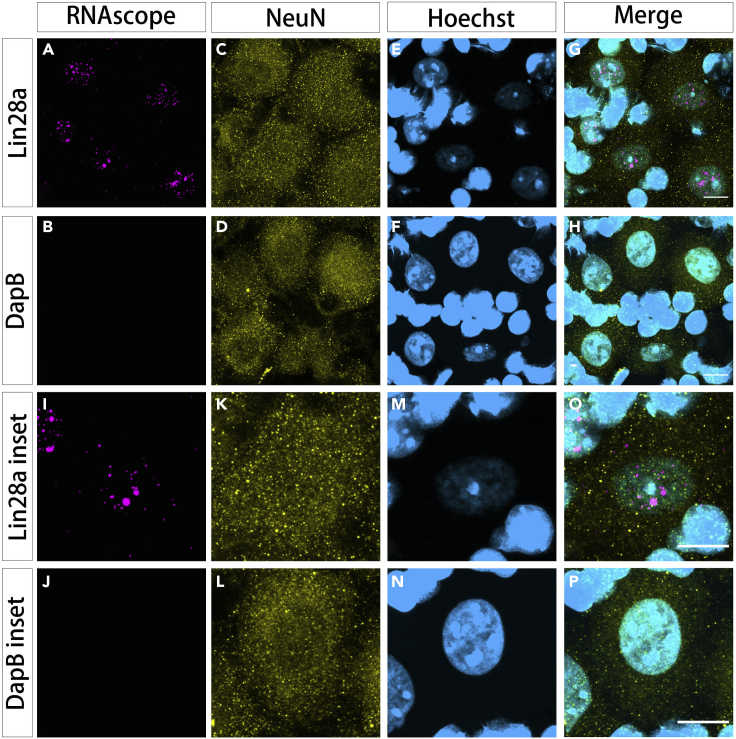
Figure 5.
Lin28a mRNA smFISH in mouse dorsal root ganglia (DRG) tissue combined with immunostaining for neuronal cell bodies and nuclei
(A and I) RNAscope channel for Lin28a mRNAs; probes were visualized using opal 570 dye [ex/em: 561 / 570–650 nm; laser intensity: 0.25%; pinhole: 1 airy unit; detector: Airyscan; detector gain: 750 V].
(B and J) RNAscope channel for negative control DapB mRNAs probe detected with opal 570 and the same confocal settings as in (A).
(C, D, K, and L) Immunohistochemistry for NeuN to mark neuron cell bodies visualized with Alexa Fluor 488 [ex/em: 488 / 494–550 nm; laser intensity: 0.25%; pinhole: 1 airy unit; detector: Airyscan; detector gain: 700V].
(E, F, M, and N) Hoechst 33342 nuclear staining [ex/em 405 nm/ 400–480 nm; laser intensity: 0.5%; pinhole: 1 airy unit; detector: GaAsP; detector gain: 700V]. Hoechst 33342 staining intensity is typically weaker in neuronal nuclei, compared with satellite glial nuclei. The Hoechst channel is therefore intentionally semi-saturated to enhance the visibility of neuronal nuclei.
(G, H, O, and P) Merged images for all channels: Lin28a mRNA/opal 570 (red); NeuN/Alexa Fluor 488 (green); Hoechst 33342 (blue). NeuN transparency is increased for visualization purposes in merged images. Scale bars: 10 μm.