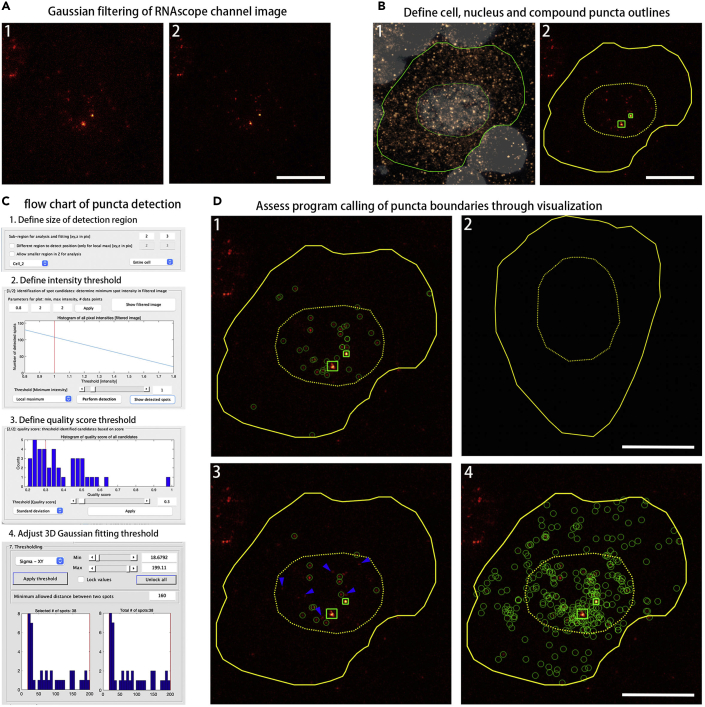
Figure 7.
Quantification of discrete mRNA puncta in DRG neurons using FISH-quant
(A) Gaussian filtering of RNAscope channel image (red). (1) Unfiltered Lin28a RNAscope channel image. (2) Lin28a RNAscope image with two-step Gaussian filtering with default Gaussian kernels. First filter: kernel = 5 (pixel) for XY and Z; second filter: kernel = 1 (pixel) for XY and Z.
(B) Define cell, nucleus and compound puncta outlines. (1) Manually delineate the outlines for neuronal cells using the NeuN immunohistochemistry channel (copper) and the nuclear outlines using Hoechst 333542 staining (bone). Define cell and nuclear outlines with polygons (yellow). Save outlines as a text file containing XY position information to a specified directory. (2) Import the cell & nuclear outline text file from the respective channel to function as an overlay in the RNAscope channel image (FQ menu > Load > Outline from other channel), and manually draw the outlines for compound puncta (if present) on RNAscope channel images. Compound puncta outlines may be defined with rectangles (green).
(C) Flow chart of puncta detection. (1) Define size of detection region (sub-region) around puncta. The region should be large enough to capture entire signal but not so large that background gets dominant. (2) Define intensity threshold for local maximum detection of puncta. (3) Define quality score threshold for detected puncta, quality scores are determined by standard deviation of all pixel intensities in the sub-region around each detected punctum. (4) Adjust 3D Gaussian fitting threshold to exclude false-positive puncta.
(D) Assessing program calling of puncta boundaries through visualization. (1) Lin28a puncta calling (green) with optimal parameters for detection [size of detection region: XY=2, Z=3; intensity threshold = 1; quality score threshold = 0.2; Gaussian fitting threshold: sigma-XY (size in XY) = 18.68–199.11; sigma-Z = 34.70–1749.72; amplitude = 2.31–403.93]. (2) DapB (negative control probe) puncta calling with the same detection settings as in (1). (3) An example appearance of inappropriate puncta calling when intensity threshold is assigned too high, missing puncta are marked with arrowheads (blue). [size of detection region: XY=2, Z=3; intensity threshold = 1.7; quality score threshold = 0; Gaussian fitting threshold: sigma-XY = 19.15–106.82; sigma-Z = 62.26–1211.90; amplitude = 4.06–204.88]. (4) An example appearance of inappropriate excess puncta calling when detection region around puncta and intensity threshold is assigned too low [size of detection region: XY=2, Z=2; intensity threshold = 0.5; quality score threshold = 0; Gaussian fitting threshold: sigma-XY = 14.92–216.18; sigma-Z = 25.44–1991; amplitude = 0–705844.00]. Scale bars: 10 μm.