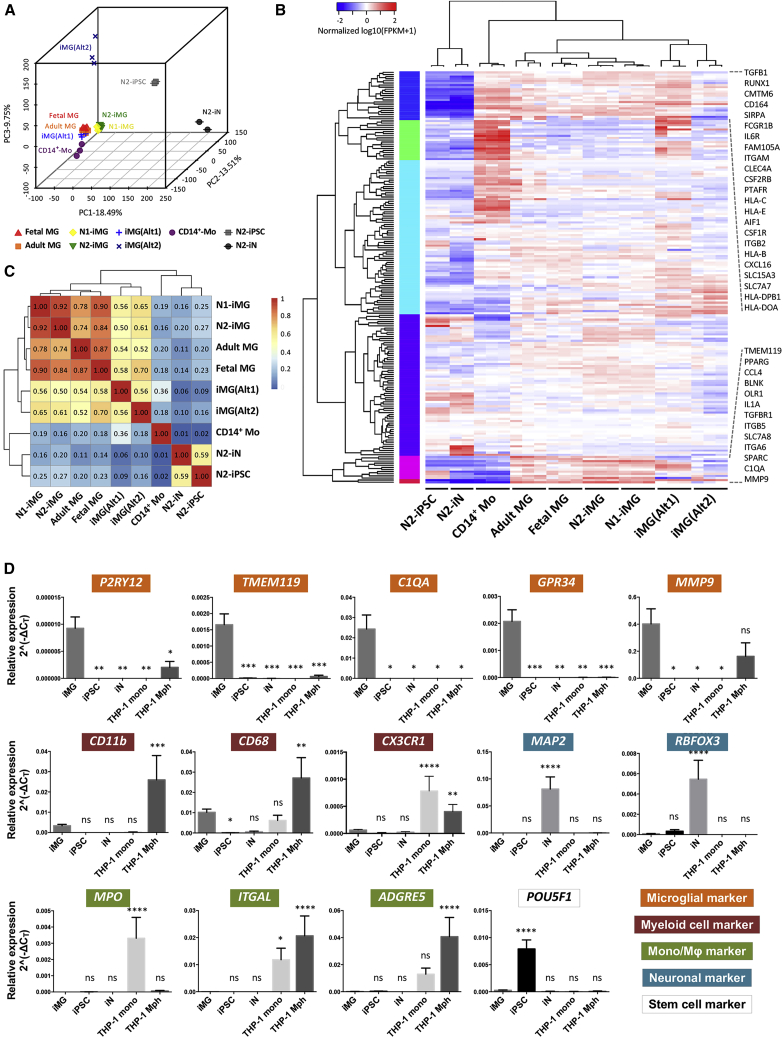
Figure 3.
iMG cells express consensus microglial markers
(A) Three-dimensional principal component analysis of N1- and N2-iMG cells (yellow and green, respectively) by whole-transcriptome sequencing of protein coding genes. The profiles of hiPSC (N2-iPSC), iN (N2-iN), and iMG cells derived from two hiPSC lines with different genetic backgrounds, N1-iMG and N2-iMG, were merged with the dataset from Abud et al. (2017) and Brownjohn et al. (2018), including cultured human primary microglia from adult and fetal microglia, iMG cells from two different methods (iMG(Alt1) and iMG(Alt2)), and CD14+ peripheral blood monocytes. Each spot represents one independently differentiated cell batch and each cell type is coded (different colors and shapes).
(B) Heatmap of 195 microglial, myeloid, and other immune-related genes. A pseudo-color is used to present the log10-transformed FPKM values (FPKM+1).
(C) Spearman correlation matrix for correlations between different cell DeSeq2 rlog-transformed raw counts of genes used in (B). Median rlog gene counts of the biological replicates were used as input. The color shows the strength and direction of the correlation.
(D) Expression of key microglial markers (P2RY12, TMEM119, C1QA, GPR34, MMP9), myeloid cell markers (CD11b, CD68, CX3CR1), monocyte and macrophage markers (MPO, ITGAL, ADGRE5), neuronal markers (MAP2, RBFOX3), and a stem cell marker (POU5F1) in N2-iMG cells after 9–12 days of induction (obtained by qRT-PCR). Fold change was calculated using the ΔCT method with RPL13A as an endogenous control. Data are means ± SEM (n = 3–11 independent experiments). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant. All compared with the iMG sample by one-way ANOVA with Fisher's least-significant difference (LSD) multiple comparisons.