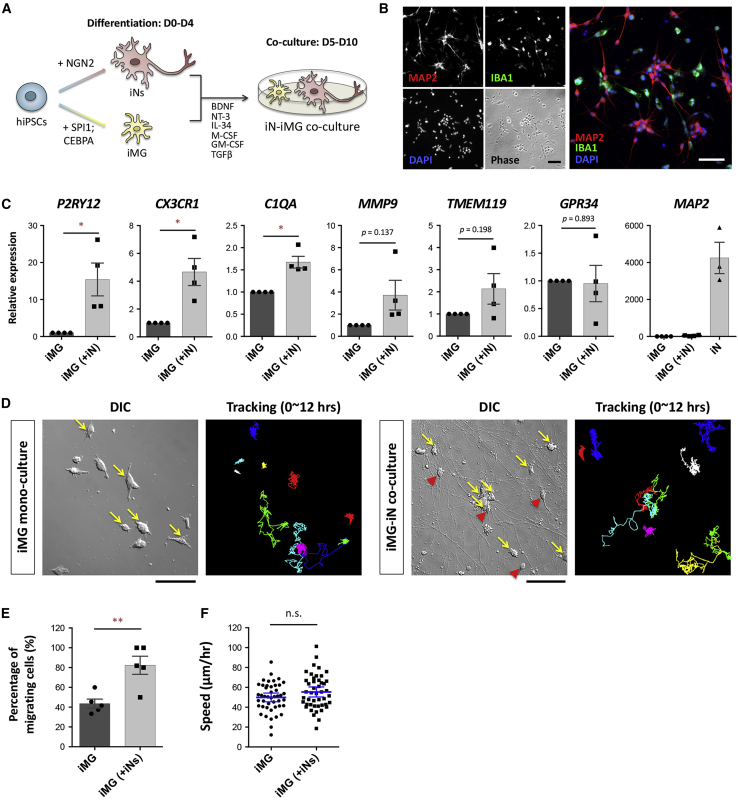
Figure 6.
Co-culture of iMG cells with hiPSC-derived neurons promotes iMG maturation
(A) Schematic overview of the co-culture protocol for iN and iMG cells generated from the same hiPSC line. The hiPSCs carrying the NGN2 or SPI1/CEBPA transgene were induced to form neurons (iNs) or microglia (iMG) separately. Next, the iN and iMG cells were co-cultured under compatible conditions from day 5 to day 10.
(B) Representative images of the N2-iN and N2-iMG co-culture immunolabeled for MAP2 (red) and IBA1 (green) at day 10. Cell nuclei are stained with DAPI (blue). Scale bar, 50 μm.
(C) Expression levels by qRT-PCR of key microglial markers (P2RY12, CX3CR1, C1QA, MMP9, TMEM119, GPR34) and the somatodendritic marker MAP2 in mono-cultured or co-cultured N2-iMG cells at 9–12 days of induction. Fold changes in target genes were calculated using the 2ˆ(-ΔΔCT) method with RPL13A as an endogenous control and relative to the expression levels found in mono-cultured iMGs. Data are means ± SEM (n = 4 independent experiments). ∗p < 0.05 by paired t test.
(D) An example of time-lapse differential interference contrast (DIC) imaging of N2-iMG cultures with or without N2-iNs (recorded for 12 h). Yellow arrows in the DIC images mark the migrating iMG cells, defined as cells with a displacement length of over two cell bodies between two continuous frames over 12 h. Red arrowheads indicate iNs. The right-side images show the cell trace results and are presented as the color-coded trajectories of each cell over 12 h. Scale bar, 100 μm.
(E) Percentage of migrating N2-iMG cells in mono-culture or in co-culture (12-h recording period). Data are means ± SEM (n = 5 independent fields in each group). ∗∗p < 0.01 by unpaired t test.
(F) The speed of migration of the N2-iMG cells in the mono-culture or co-culture (12-h recording period). Data are means ± SEM (n = 45 and 46 cells from 5 fields in the N2-iMG and N2-iMG(+N2-iN) groups, respectively). n.s., not significant by unpaired t test.