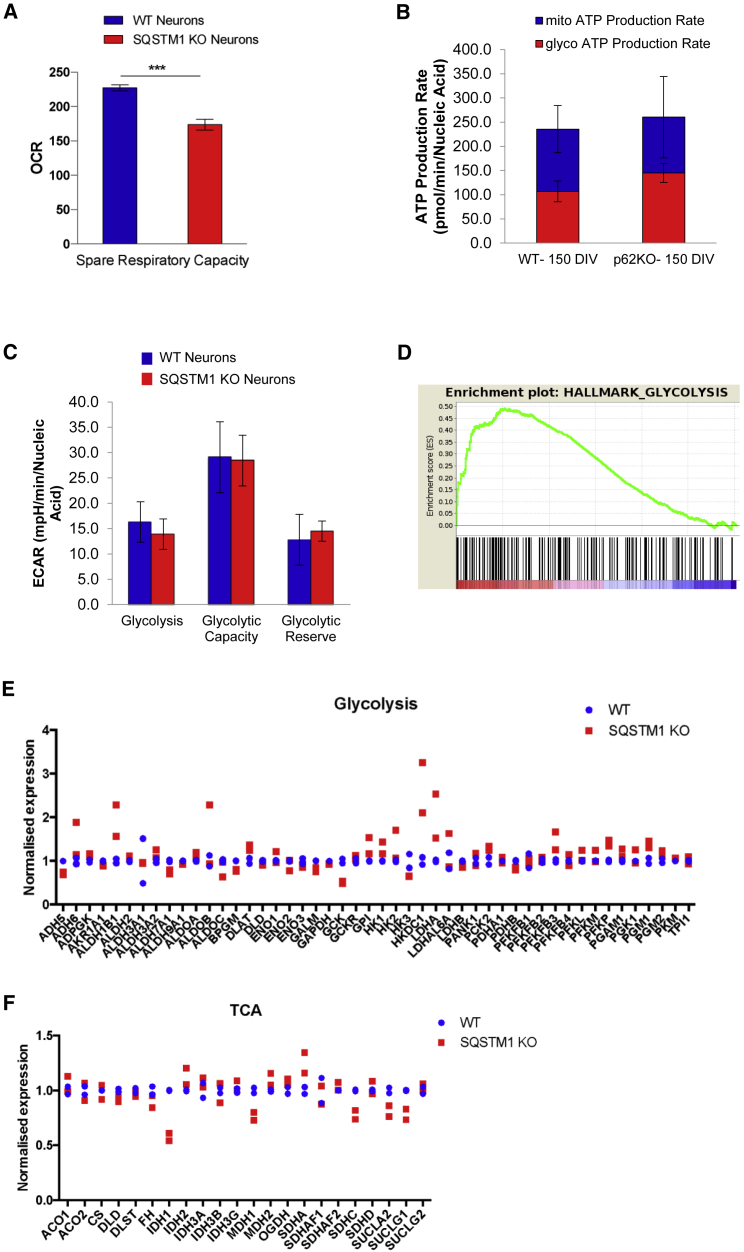
Figure 3.
Deletion of SQSTM1 has small but significant effect on mitochondrial spare respiratory capacity
(A) Results of spare respiratory capacity from Seahorse XF Mito Stress Test performed on cortical neurons derived from SQSTM1 KO and WT iPSCs (RBi001-A). Data presented as mean ± SEM of three independent experiments and statistical differences were tested by Welch's t test. ∗∗∗p < 0.001.
(B) Seahorse XF Real-Time ATP Rate Assay in iPSC-derived neurons with or without SQSTM1. Basal ATP production rates from mitochondrial (blue) and glycolytic (red) pathways were quantified and are summarized in the stacked bar chart. Data are presented as the mean ± SEM for eight technical replicates, and statistical differences were tested by Welch's t test.
(C) Results of Seahorse XF Glycolysis Stress Test from cortical neurons derived with or without SQSTM1 KO. Individual parameters of glycolytic function including basal glycolysis, glycolytic capacity, and glycolytic reserve, were obtained from the extracellular acidification rates (ECAR). Data are presented as mean ± SEM for six technical replicates, and statistical differences were tested by Welch's t test.
(D) GSEA enrichment plot of the glycolysis pathway. The x axis is genes (vertical black lines) and y axis represents enrichment score (ES), which represents the enrichment of the glycolysis pathway at the top of the ranked gene list. The colored bar at the bottom represents the degree of correlation of genes with the SQSTM1 KO phenotype (red for positive and blue for negative correlation).
(E) Relative mRNA expression of glycolytic genes in neurons derived from SQSTM1 KO and WT iPSCs (RBi001-A). Dots indicate mean datapoints from each of two independent neural differentiations for each line (SQSTM1 KO and WT).
(F) Relative mRNA expression of TCA genes in neurons derived from SQSTM1 KO and WT iPSCs (RBi001-A). Dots indicate mean datapoints from each of two independent neural differentiations for each line (SQSTM1 KO and WT).