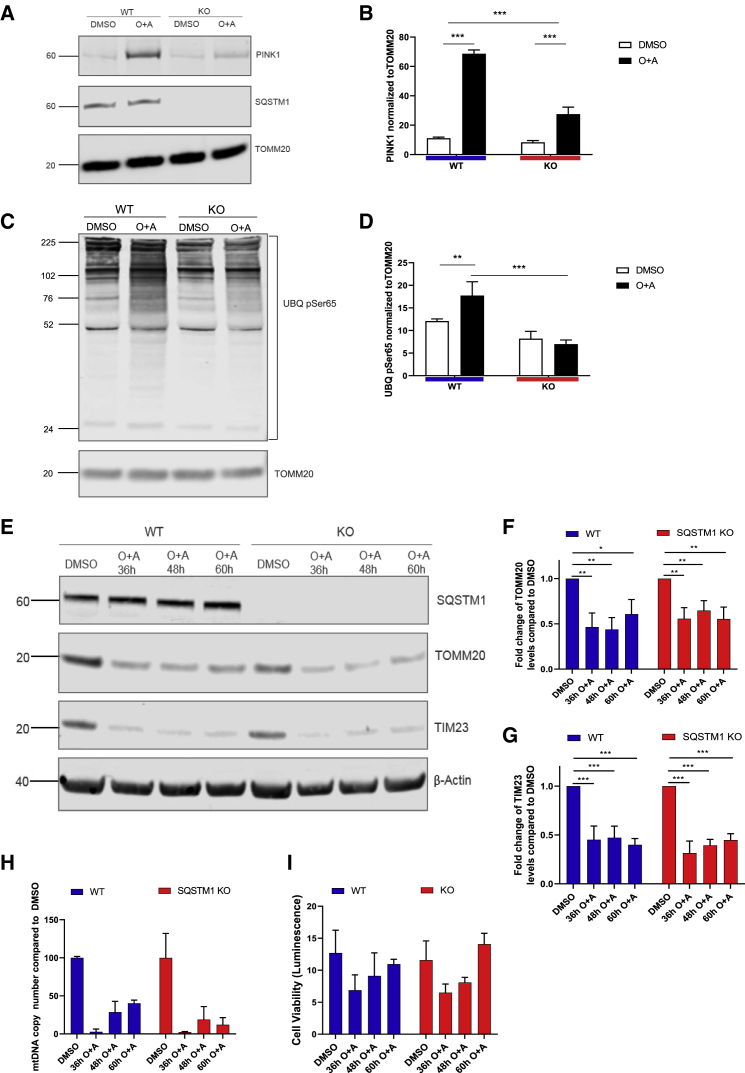
Figure 4.
SQSTM1 is not required for mitophagy in iPSC-derived cortical neurons
(A) Immunoblots probed for mitochondrial components PINK1, SQSTM1, and TOMM20 after cells were treated with 1 μM oligomycin and antimycin A.
(B) Bar graph displays quantified images showing PINK1 normalized to TOMM20 (loading control). Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by two-tailed t test. ∗∗∗p < 0.001.
(C) Immunoblot for Ser65 (UBQ pSer65) and TOMM20 (using the same samples as examined in A).
(D) Bar graph displays quantified images showing UBQ pSer65 normalized to TOMM20 (loading control). Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by two-tailed t test. ∗∗∗p < 0.001.
(E) Immunoblot for TOMM20, TIM23, SQSTM1, and β-actin from neurons treated for 36–60 h with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control).
(F and G) Bar graphs display quantified images showing fold change versus DMSO control of TOMM20 and TIM23 versus β-actin loading control. Data are expressed as mean ± SEM of three independent experiments. Statistical differences were tested by two-tailed t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(H) Quantitative mitochondrial DNA copy number in neurons, treated for 36–60 h with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control), as determined by real-time PCR. Data are expressed as the mean ratio of mitochondrial-to-nuclear genes ± SEM of three technical replicates.
(I) Cellular viability in neurons, treated for 36–60 h with 1 μM oligomycin and 1 μM antimycin A (or DMSO as control), analyzed luminometrically using the RealTime-Glo MT Cell Viability Assay. Data are expressed as mean ± SEM of three technical replicates.