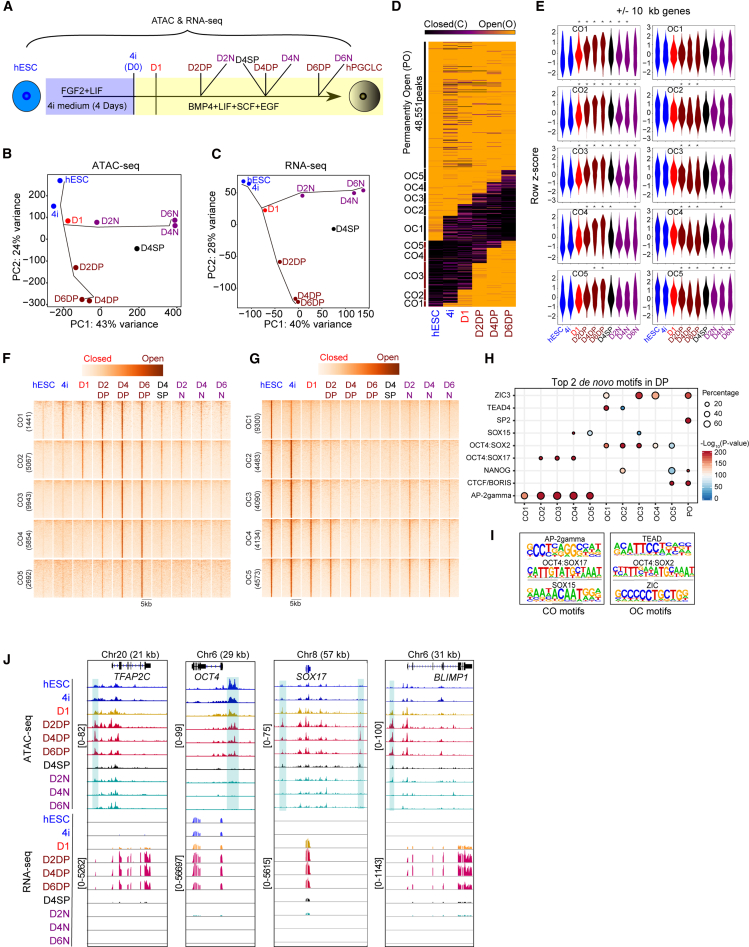
Figure 1.
Chromatin accessibility and gene regulation dynamics during hPGCLC induction
(A) Schematic representation of time course ATAC-seq and RNA-seq library induction during the hPGCLC induction from hESCs. Day is represented as “D,” EpCAM+/INTEGRINα6+ cells are represented as DP, EpCAM−/INTEGRINα6− cells are represented as N, and EpCAM+/INTEGRINα6− cells are represented as SP.
(B and C) PCA of ATAC-seq (B) and RNA-seq data (C). Cell types are labeled as described in (A) and two independent replicates are merged.
(D) Dynamically closed-to-open (CO), open-to-closed (OC), and permanently open (PO) chromatin regions are clustered and shown as a heatmap. CO, OC, and PO refer to closed in hESCs but open in D6DP, open in hESCs but closed in D6DP, and PO in both hESCs and D6DP, respectively.
(E) Violin plots showing the expression levels of all genes with a TSS within 10 kb of an ATAC-seq peak for each CO or OC group. The Wilcoxon rank-sum test was performed. ∗p < 0.01.
(F and G) Heatmap showing the genome coverage of ATAC-seq signals on each CO (F) and OC (G) group.
(H) Bubble plot showing the top 2 de novo motifs in COs/OCs.
(I) Selected top ranked de novo motifs from CO (left) and OC (right).
(J) Representative genome coverage plots for ATAC-seq and RNA-seq signals for key germ cell genes.