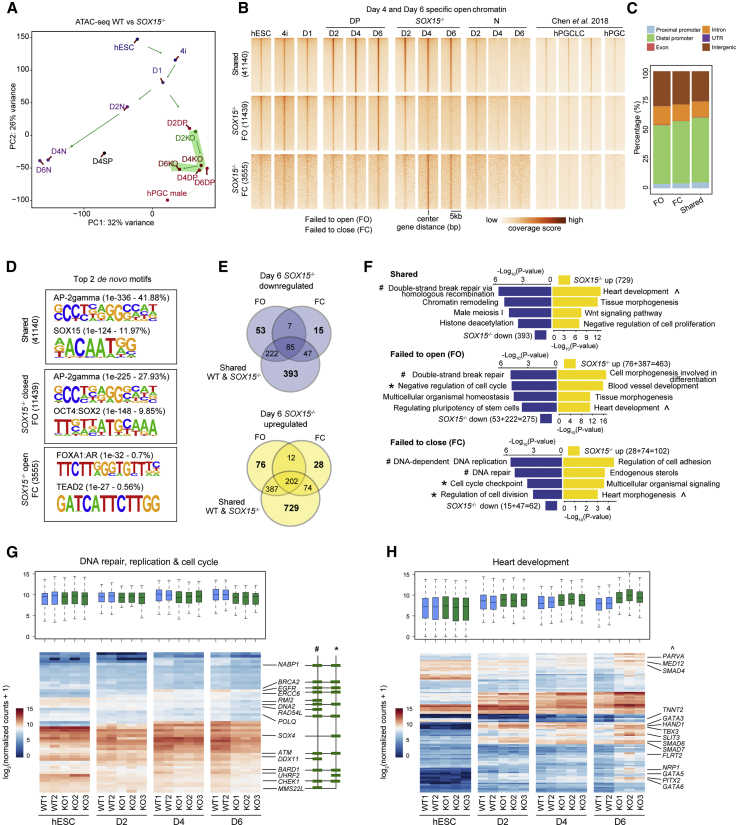
Figure 5.
The suppression of somatic gene expression mediated by SOX15 is associated with chromatin accessibility
(A) PCA plot showing ATAC-seq analysis of the hPGCLC induction under the normal and SOX15 KO states. Two independent replicates are merged.
(B) Heatmap of ATAC-seq signals in the indicated samples over shared-open chromatin regions constituting 41,140 peaks, SOX15−/− FO regions constituting 11,439 peaks, and SOX15−/− FC regions constituting 3,555 peaks.
(C) Bar plot showing the percentage of genomic features from FO, FC, and shared regions.
D) Top 2 de novo motifs from shared, FO, and FC genomic regions. The name of the motifs with respective p value and percentage are shown on each motif.
(E) Venn diagram showing the intersection of nearby genes from FO, FC, or shared regions that shared with the downregulated and upregulated genes in day 6 SOX15−/− cells, respectively. Log2 fold change > 0.5.
(F) GO analysis for upregulated and downregulated genes nearby shared, FO, and FO regions, respectively, as shown in (E).
(G and H) Boxplots (with the median and 25th and 75th percentiles) and heatmap showing the expression patterns of specific genes representing the GO terms of DNA repair, DNA replication, and cell cycle (G), or heart development (H). The symbols #, ∗, and ˆ represent the GO terms shown in (F) and key genes are indicated.