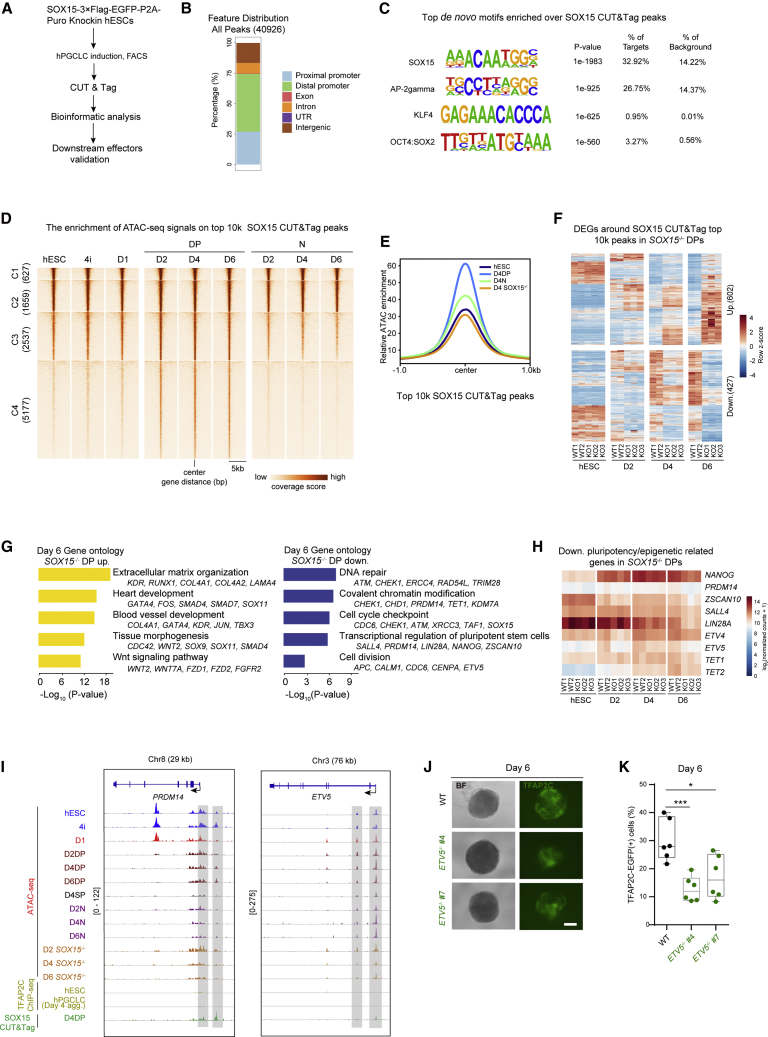
Figure 6.
SOX15 exerts its function in hPGCLC maintenance by directly suppressing somatic gene expression and sustaining latent pluripotency
(A) Schematic representation of the SOX15 CUT&Tag analysis workflow in hPGCLCs.
(B) Bar plot showing the percentage of genomic feature distribution of SOX15 peaks.
(C) The top binding motifs enriched in SOX15 peaks.
(D) Heatmap of ATAC-seq signals in the indicated samples over the top 10k SOX15 peaks.
(E) Pileup of the ATAC-seq signals over the top 10k SOX15 peaks regions in the indicated cells.
(F) Heatmap showing the expression patterns of upregulated or downregulated genes around the top 10k SOX15 peaks in day 6 SOX15 KO DP cells.
(G) GO analysis for the upregulated or downregulated genes as described in (F).
(H) Heatmap showing the expression patterns of downregulated pluripotency-related genes in SOX15 KO DP cells.
(I) Selected genomic views showing the ATAC-seq signals, TFAP2C ChIP signals (Chen et al., 2019), and SOX15 signals at the PRDM14 and ETV5 genome loci in the indicated samples. The specific open regions with SOX15 signals and decreased ATAC-seq signals from day 4 KO DP cells compared with those in DP cells are marked with a gray box.
(J) Bright-field (BF) and fluorescence (TFAP2C-EGFP) images of floating embryoids from WT and ETV5−/− lines at day 6. Scale bar, 200 μm.
(K) The percentages of TFAP2C-EGFP(+) cells of floating embryoids from day 6 WT (black) and ETV5 KO lines (green) upon hPGCLC induction via the 4i method. Results of six independent experiments are shown (n = 6). Two-tailed Student's t test was performed, ∗p < 0.05, ∗∗∗p < 0.001.