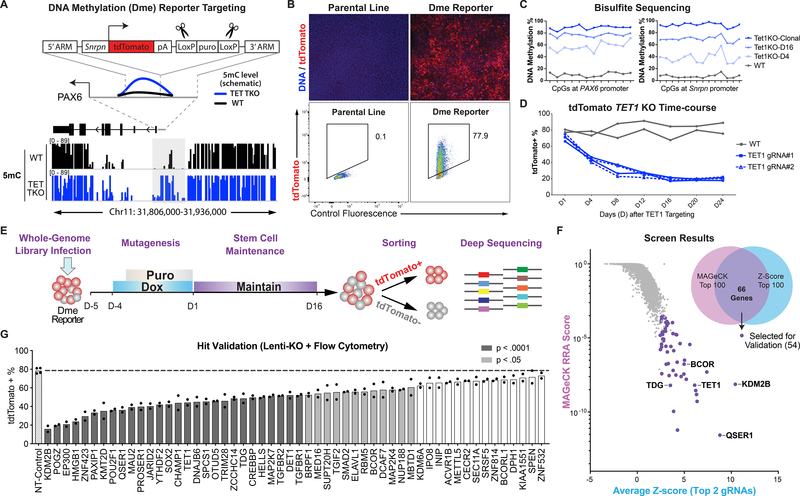
Fig. 1.
A genome-wide CRISPR/Cas screen for regulators of DNA Methylation. (A) Knockin strategy for generating iCas9;PAX6-DmetdTom reporter in reference to DNA methylation at PAX6 locus from Whole Genome Bisulfite Sequencing in WT (black) and TET TKO (blue) hESCs (14). LoxP-flanked puromycin was excised with transient expression of Cre recombinase. (B) Immunofluorescence staining (with anti-RFP antibody) and flow cytometry for tdTomato expression. (C) Bisulfite sequencing of PAX6 promoter and inserted Snrpn promoter. TET1KO-D4 and D16 denote KO using lentivirus expressing TET1_gRNA#1 where cells were collected on day 4 (D4) or day 16 (D16). Day 1 (D1) denotes 5 days after doxycycline treatment. (D) Summary of flow cytometry of lentivirus TET1 KO cells. (E) Screen schematic: doxycycline induction for Cas9 expression and puromycin selection for integration of lentiviruses. (F) Screen results: MAGeCK RRA score vs. average of two highest Z-scores for each gene. Venn diagram indicates overlap between top 100 genes in each ranking method. Genes selected for validation and selected top hits are highlighted. (G) Summary of flow cytometry of 54 lines expressing gRNAs targeting genes for validation compared to non-targeting control line (n=2). P values = one-way ANOVA followed by Dunnet multiple comparisons test.