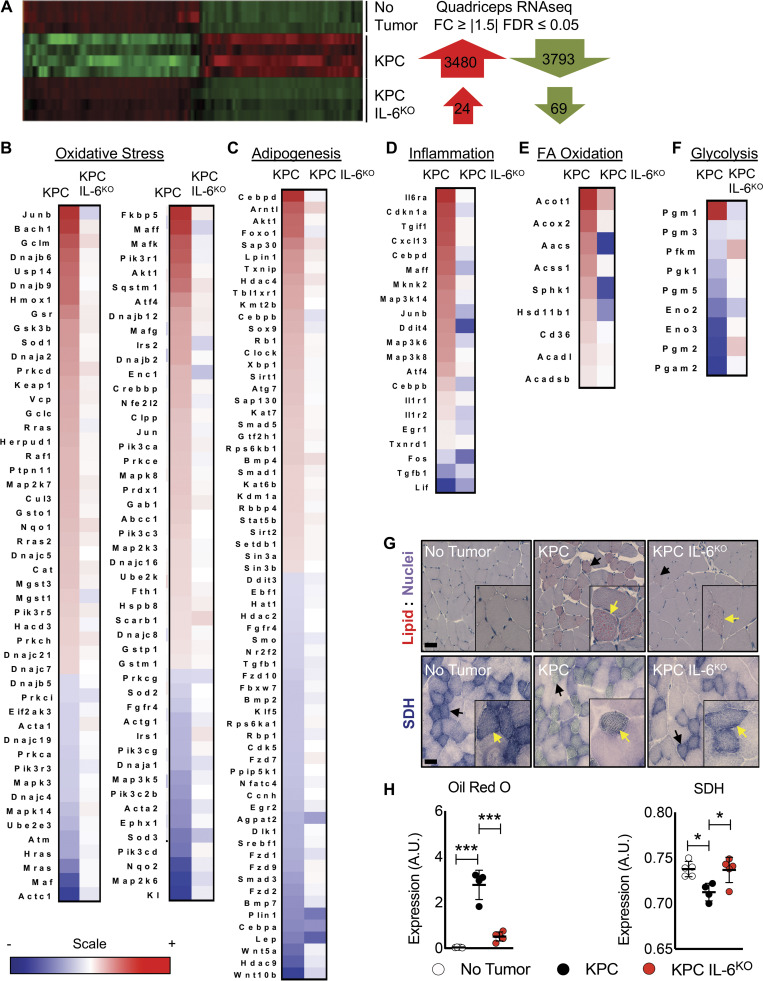
Figure 5.
Deletion of IL-6 from KPC cells reduces activation of key cachexia pathways in muscle. (A) Isolated RNA from the quadriceps of no tumor (n = 3), KPC tumor (n = 4), and KPC IL6KO tumor (n = 4) mice was sequenced, and differentially regulated genes (FC ≥ |1.5| and FDR ≤ 0.05) were compared across groups. (B–F) Ingenuity Pathway Analysis using quadriceps RNA-seq data identified various altered pathways and their associated genes (shown in heat map format) that have roles in muscle wasting, including oxidative stress (B), adipogenesis (C), inflammation (D), FA oxidation (E), and glycolysis (F). The scale bar illustrates increased (red) and decreased (blue) gene expression for the heat maps. (G and H) Measurements of muscle lipids using ORO staining (G, top, and H, left) and SDH activity as a marker for mitochondria oxidative capacity (G, bottom, and H, right) were performed on quadriceps muscle cross sections (n = 5 per group). Scale bar = 50 µm; black arrows in G indicate fibers with increased lipid accumulation and aberrant SDH reactivity at lower magnification; yellow arrows in G indicate fibers with increased lipid accumulation and aberrant SDH reactivity in higher magnification insets. Error bars represent mean and SD, and significant differences (*, P < 0.05; ***, P < 0.0001) were determined using ANOVA and Tukey’s multiple comparisons test.