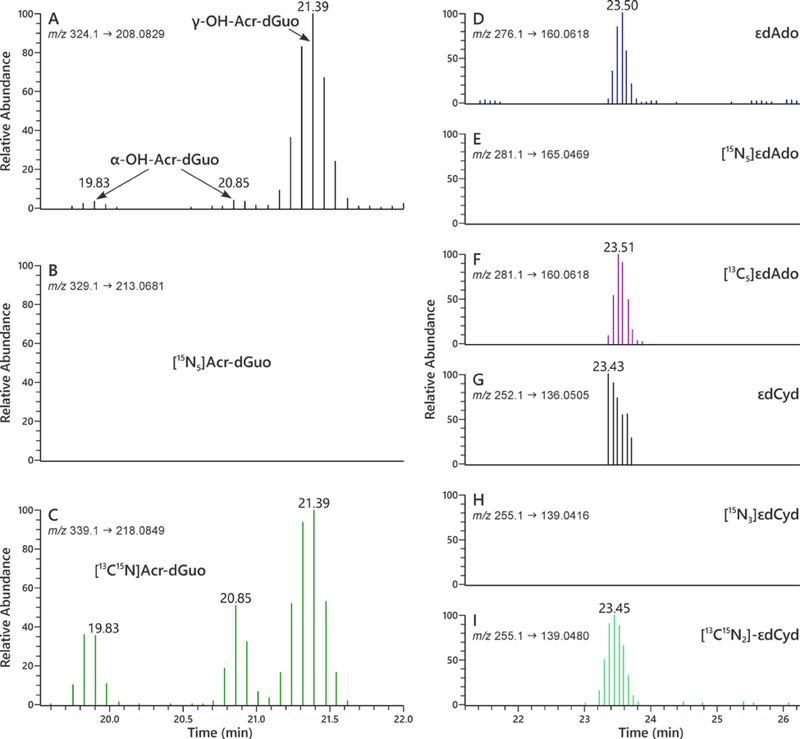
Figure 4.
LC-NSI-HRMS/MS chromatograms obtained upon analysis of human oral cell DNA obtained from an oral rinse sample for Acr-dGuo (1 and 2), εdAdo (3), and εdCyd (4). SRM was carried out at (A) m/z 324.1 → m/z 208.0829 for adducts 1 and 2, (B) m/z 329.1 → m/z 213.0681 for spiked 15N bacterial DNA adducts [15N5]1 and 2, and (C) m/z 339.1 → m/z 218.0849 for internal standard adducts [13C1015N5]1 and 2; SRM was carried out at (D) m/z 276.1 → m/z 160.0618 for adduct 3, (E) m/z 281.1 → m/z 165.0469 for spiked 15N bacterial DNA adduct [15N5]3, and (F) m/z 281.1 → m/z 160.0618 for internal standard adduct [13C5]3; SRM was carried out at (G) m/z 252.1 → m/z 136.0505 for adduct 4, (H) m/z 255.1 → m/z 139.0416 for spiked 15N bacterial DNA adduct [15N3]4, and (I) m/z 255.1 → m/z 139.0480 for internal standard adduct [13C15N2]4. In each case, the transition monitored represents loss of the deoxyribose ring with transfer of a proton to the base, e.g. ([M + H]+ → [BH]+).