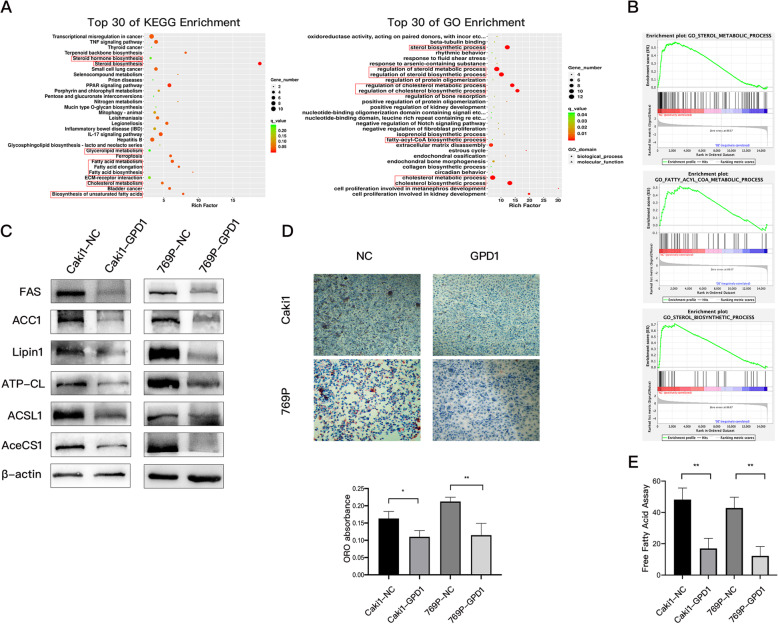
Fig. 6.
Overexpression of GPD1 inhibited lipid metabolism in ccRCC cells. (A) KEGG analysis and gene ontology (GO) analysis of the differentially expressed genes (|log2FC| > 1, FDR-adjusted p < 0.05) in 786O-NC and 786O-GPD1 cells. (B) GSEA revealed that sterol biosynthetic gene sets, fatty acyl-CoA metabolism gene sets, and sterol metabolism gene sets were less enriched in 786O-GPD1 cells (FDR < 25% and p < 0.05). (C) Western blotting of indicated proteins in Caki1-NC, Caki1-GPD1, 769P-NC and 769P-GPD1 cells. (D) Images and quantifications (at 560 nm wavelength) of Oil Red O staining in Caki1-NC, Caki1-GPD1, 769P-NC, and 769P-GPD1 cells. (E) Quantitation of free fatty acids in Caki1-NC, Caki1-GPD1, 769P-NC, and 769P-GPD1 cells. Data are shown as the mean + SD (n = 3). Statistical analysis was performed using an unpaired Student’s t test with a two-tailed distribution (* p < 0.05, **p < 0.01). Abbreviations: GPD1, glycerol-3-phosphate dehydrogenase 1; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto encyclopedia of genes and genomes; ccRCC, clear cell renal cell carcinoma; CCK-8, cell counting kit