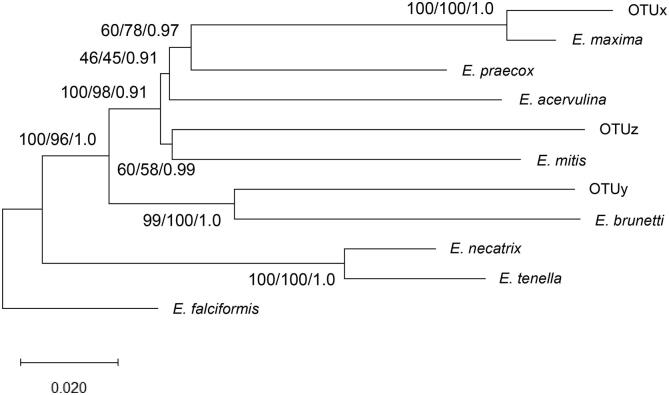
Fig. 6.
Optimal Neighbour-Joining tree inferred using a 4135 amino acid alignment of 56 concatenated Eimeria gene models rooted on Eimeria falciformis. Support for each node is presented, indicating outcomes from Maximum Likelihood (ML)/Neighbour-Joining (NJ)/Bayesian methods. For ML, the General Time Reversible (GTR) model was used with a gamma distribution. For NJ evolutionary distances were calculated using the Kimura 2-parameter model. For Bayesian inference analysis the GTR+G model was used, including four runs with 1,000,000 generations, a sample frequency of 10, and 25% burn-in.