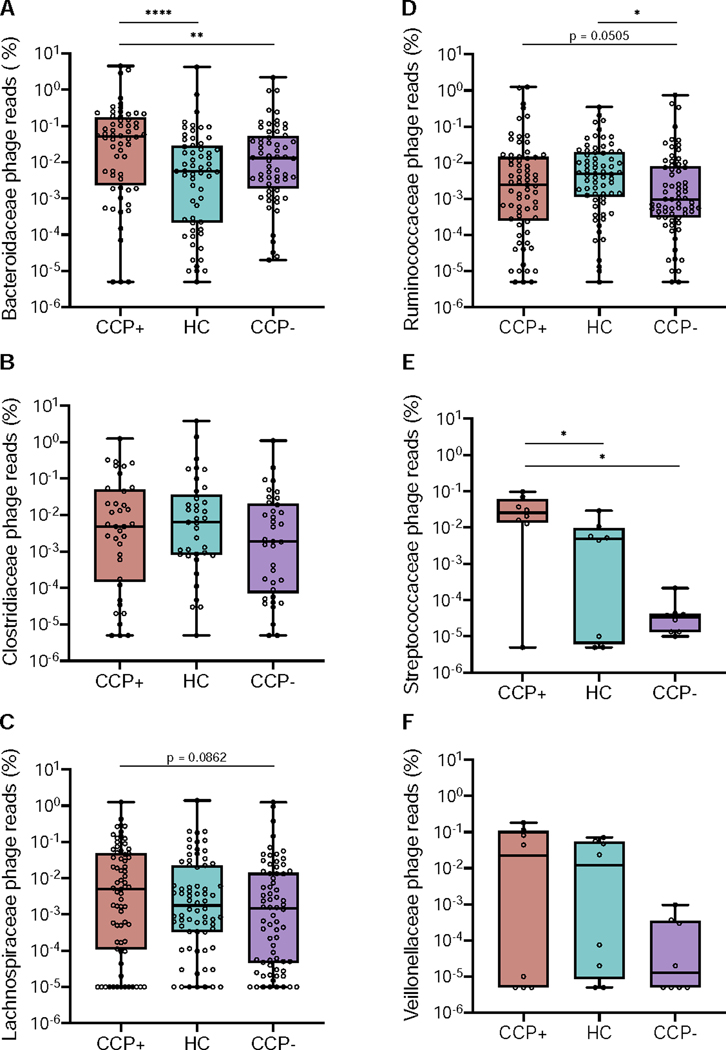
Figure 3.
Phage-host assignments for curated VLP contigs reveal cohort-based differential read recruitment among several bacterial families. Relative abundances were calculated for all VLP reads mapped to phages predicted to target Bacteroidaceae (A), Clostridiaceae (B), Lachnospiraceae (C), Ruminococcaceae (D), Streptococcaceae (E), and Veillonellaceae (F) bacterial families. Scaffold abundances were averaged across all samples and statistics were determined by nonparametric Wilcoxon tests (* p < 0.05, ** p < 0.01, **** p < 0.0001).