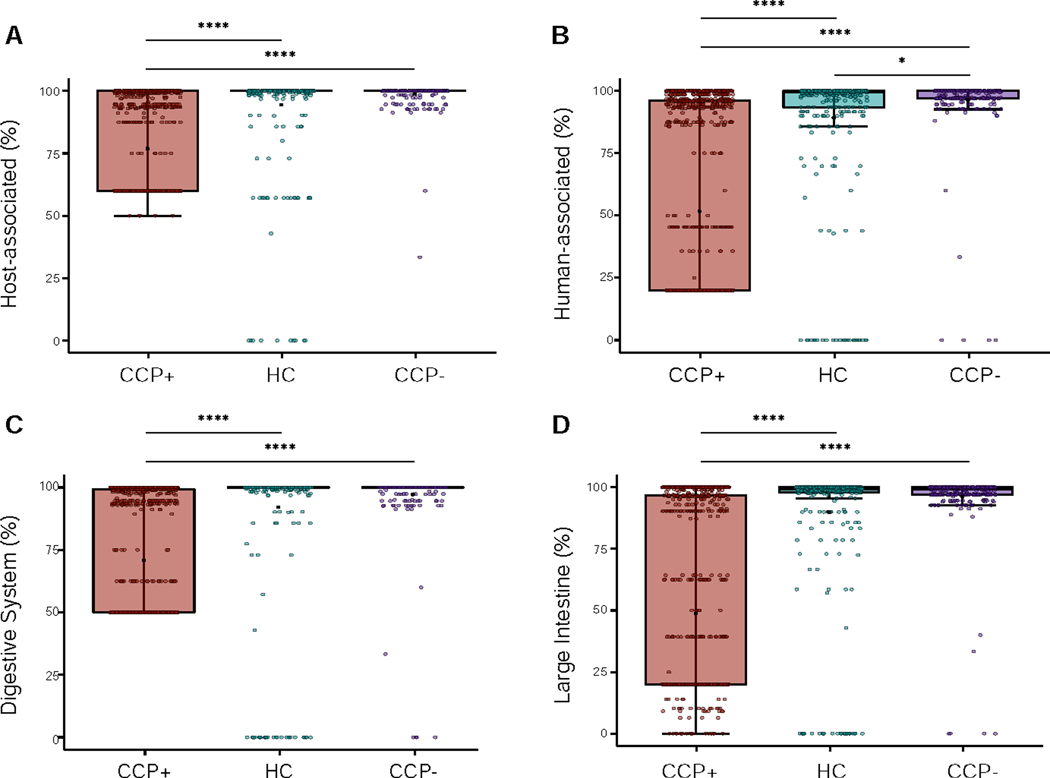
Figure 4.
CRISPR spacer host metadata reveal CCP+ phages represent greater variability in microbial host ecology. (A) JGI/GOLD Ecosystem Distribution showing the percent host-associated spacers calculated for each contig based on cohort distribution. (B) Ecosystem Category distribution showing the percent human-associated spacers. (C) Ecosystem Type distribution showing the percent of contigs that contain spacers originating from the digestive system. (D) Ecosystem Subtype showing the percent of contigs that contain spacers originating from the large intestine microenvironment. Statistical significance was determined using pairwise Wilcoxon rank sum tests for comparisons between the three groups, using the Benjamini-Hochberg correction for multiple testing comparisons (* p = 0.023, **** p < 2 × 10−16). See also Figure S4 and Table S6.