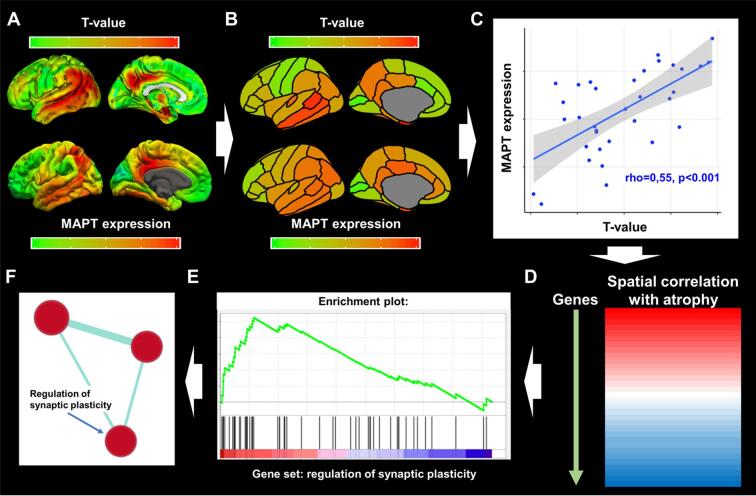
Fig. 1.
Gene-set enrichment analyses and grouping of gene sets. Panel A displays voxel-wise differences in gray matter volumes between AD subjects and cognitively normal controls (top row; T-maps, adjusted for covariates), and brain-wide gene expression values (MAPT as an example) obtained from www.meduniwien.ac.at/neuroimaging/mRNA.html (bottom row). Panel B displays the parcellation of the T-map and gene expression values into regions-of-interest (ROI) from the Desikan-Killiany atlas. Panel C displays Spearman’s correlations between T-values and gene expression values within ROIs. In this plot, each datapoint represents grey matter volume differences with controls within the 34 ROIs on the x-axis and gene-expression values in the corresponding ROI on the y-axis. The steps in panel A through C are repeated for all 18,686 genes, yielding a correlation spectrum that is ranked from positive to negative (Panel D). GSEA software then produces an enrichment score that indicates whether a gene set, as a whole, preferentially falls towards one end of the correlation spectrum (Panel E; with the gene set ‘synaptic plasticity’ as an example). Panel F displays results from the gene set grouping analysis in Cytoscape where significantly enriched gene sets are plotted as interrelated nodes connected by edges denoting their overlapping genes.