Fig 1.
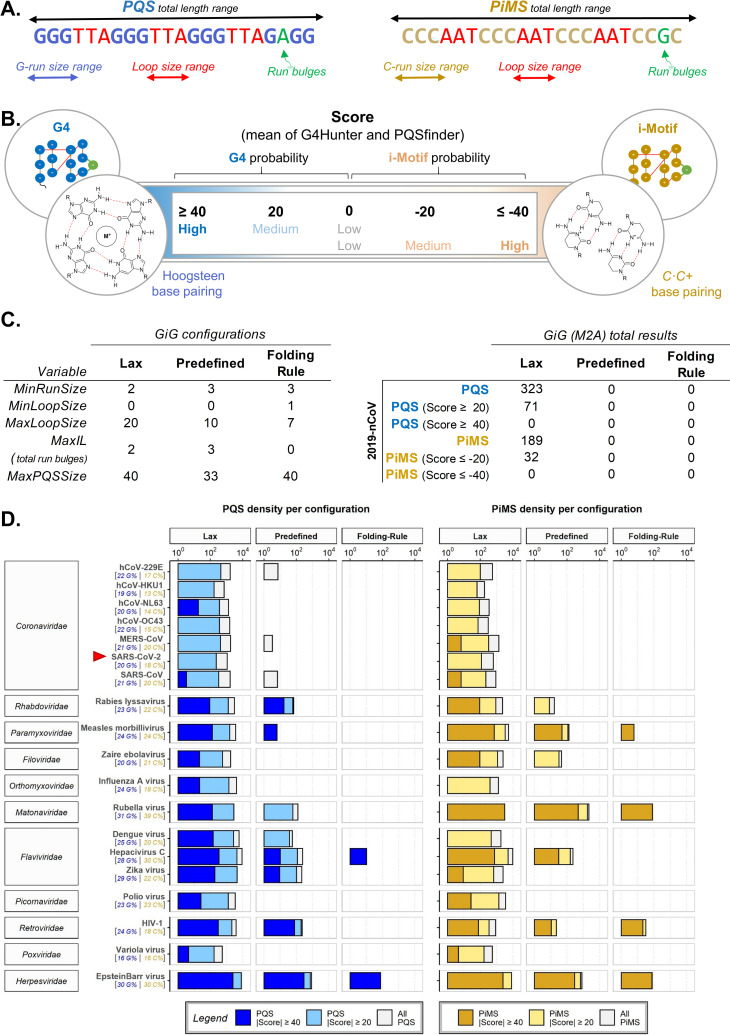
A. Results with G4-iM Grinder depend on the quadruplex definitions introduced to the algorithm. Sizes of G- or C-runs, loops and the entire sequence, together with an acceptable number of bulges within the runs are part of the definitions. B. The structures found with GiG under the definitions proposed by the user can be evaluated for their in vitro probability of formation. More positive scores mean that the sequence is more capable of forming G4s, whilst more negative values mean that it is more capable of forming iMs. C. Left, Quadruplex definitions used by GiG’s search engine in this work. C. Right, Total results found within the SARS-CoV-2 by configuration and score criteria. D. PQS and PiMS densities (per 100000 nucleotides) found per different configuration and score criteria for 19 viruses. The G and C content (as a percentage) is shown under each virus. X scale is in logarithmic scale (base 10). Results are categorized by their |score|: intense colours (blue for PQS, yellow for PiMS) are the most probable to form in vitro (|score| ≥ 40), lighter bars are the density of structures with at least a |score| ≥ 20 and grey bars are the densities without the score filter.