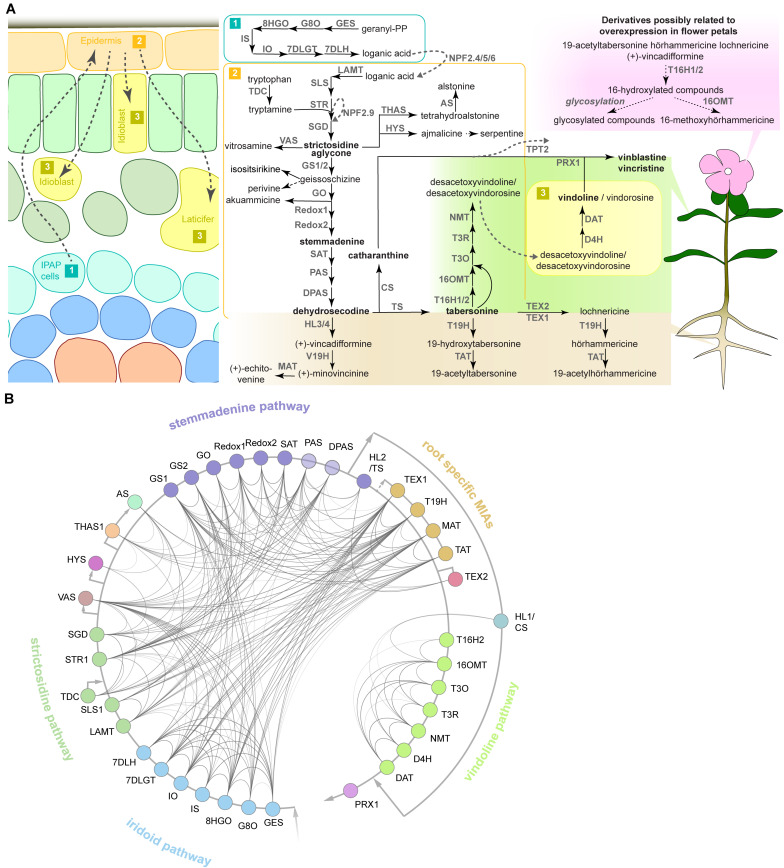
FIGURE 1.
The MIA pathway consists of different branches with corresponding co-expression networks. (A) The MIA network and known or presumed cellular and organ-specific expression of the encoding MIA biosynthesis genes based on the current knowledge and available data. MIA biosynthesis is compartmentalized into the three different cell types shown on the left, although a cell type-specific expression has not been experimentally verified for all MIA biosynthesis genes such as the root MIA biosynthesis genes. Available organ-specific transcriptome data revealed that in particular two MIA branches are either root- or leaf-specific as shown on the right. In our study, transient overexpression of TFs in C. roseus flower petals is used to assess the impact on MIA biosynthesis at the gene expression level (by qPCR) and MIA metabolite level (by LC-MS analysis). Some MIA derivatives are detected in this flower petal expression system in this study, but it is not known where they occur in wild-type plants (upper right). Arrows indicate known characterized enzymatic reactions and corresponding enzymes; dashed black arrows indicate assumed enzymatic or chemical reactions for which no specific enzyme has been identified. Dashed gray arrows indicate transport and corresponding transporters. Note that NPF2.9 is an intracellular transporter conferring vacuolar export of strictosidine and TPT2 is an epidermis-specific transporter exporting catharanthine to the leaf surface. For clarity, only selected MIA metabolites are shown. (B) Co-expression network of MIA biosynthesis genes. From an expression atlas of 82 samples KNN networks were created. A line between genes indicates co-expression. Opacity of the lines is scaled according to the mutual rank of co-expression between two genes in their KNN 500 clusters (darker = stronger co-expression). To simplify the complex MIA biosynthesis network, we have colored the different pathway branches. Abbreviations: GES, geraniol synthase; G8O, geraniol-8-oxidase; 8HGO, 8-hydroxygeraniol oxidoreductase; IS, iridoid synthase; IO, iridoid oxidase; 7DLGT, 7-deoxyloganetic acid glucosyl transferase; 7DLH, 7-deoxyloganic acid hydroxylase; LAMT, loganic acid O-methyltransferase; SLS, secologanin synthase; TDC, tryptophan decarboxylase; STR, strictosidine synthase; NPF, nitrate/peptide family; SGD, strictosidine β-glucosidase; VAS, nitrosamine synthase; THAS, tetrahydroalstonine synthase; AS, a lstonine synthase: HYS, heteroyohimbine synthase; GS, geissoschizine synthase; GO, geissoschizine oxidase; SAT, stemmadenine-O-acetyl- transferase; PAS, precondylocarpine acetate synthase; DPAS, dihydroprecondylocarpine synthase; CS, catharanthine synthase; TPT, catharanthine transporter; T16H, tabersonine 16-hydroxylase; 16OMT, 16-hydroxytabersonine O-methyltransferase; T3O, tabersonine 3-oxygenase; T3R, tabersonine 3-reductase; NMT, 2,3-dihydro-3-hydroxytabersonine-N-methyltransferase; D4H, desacetoxyvindoline 4-hydroxylase; DAT, deacetylvindoline 4-O-acetyltransferase; PRX, peroxidase; HL, hydrolase; V19H, vincadifformine-19-hydroxylase; MAT, minovincinine-19-O-acetyltransferase; T19H, tabersonine 19-hydroxylase; TAT, 19-hydroxytabersonine 19-O-acetyltransferase; TEX, tabersonine epoxidase.