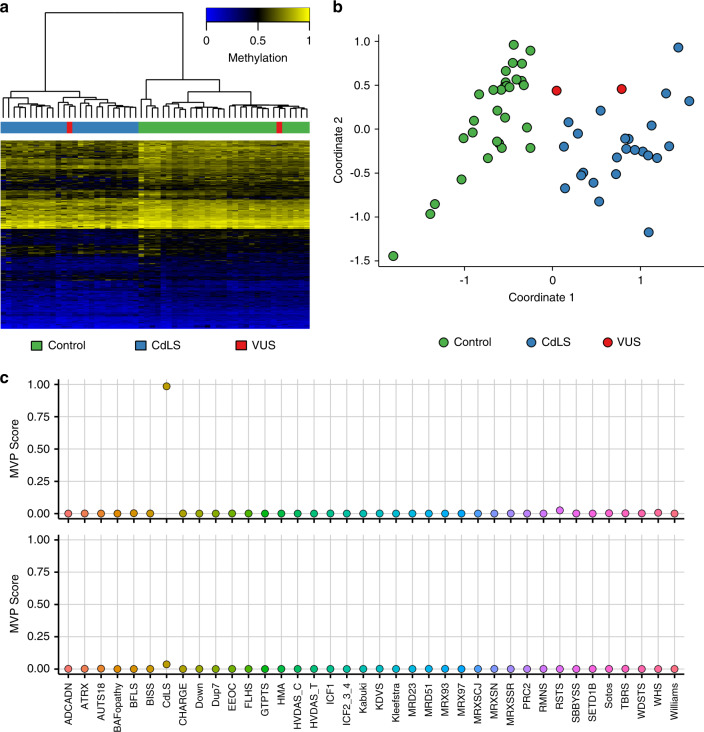
Fig. 2. DNA methylation (EpiSign) analysis of peripheral blood from two subjects with variants of unknown clinical significance (VUS) in SMC1A, the causative gene for Cornelia de Lange syndrome (CdLS) type 2.
The variants are SMC1A:c.598A>C, p.(Lys200Gln) (labeled red and clustering with CdLS samples) and SMC1A:c.1280A>G, p.(Glu427Gly) (labeled red and clustering with control samples). (a) Hierarchical clustering and (b) multidimensional scaling plot of subjects with a confirmed CdLS episignature, controls, and the VUS under investigation. The SMC1A:c.598A>C VUS clustering with the CdLS samples indicates it has a DNA methylation signature similar to that seen in other CdLS samples and suggesting that the variant is pathogenic, while SMC1A:c.1280A>G is likely benign. (c) Methylation variant pathogenicity (MVP) score, a multiclass supervised classification system capable of discerning between the 43 different episignatures in EpiSign V2, was applied to the SMC1A:c.598A>C likely pathogenic variant (top) and the SMC1A:c.1280A>G likely benign variant (bottom). This classification system generates a probability score for each episignature, with a score near 1 indicating that the sample has an episignature similar to the reference episignature.