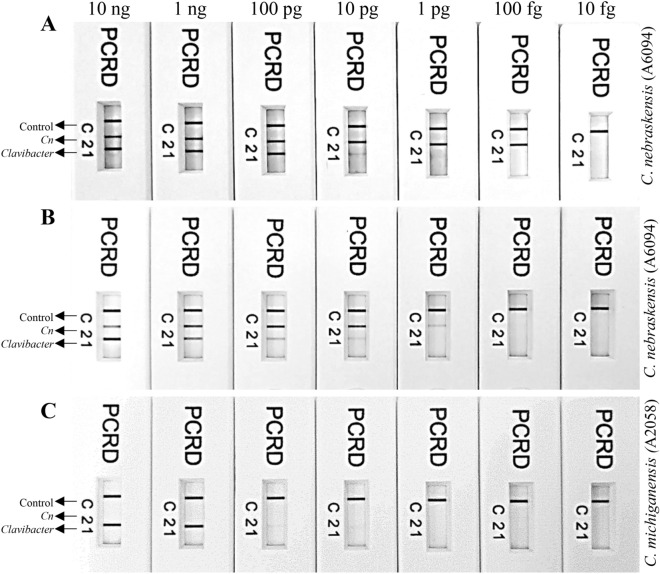
Figure 4.
Limit of detection determination of multiplex RPA assays developed for specific detection of genus Clavibacter in general and C. nebraskensis. Line 1: detects Clavibacter species/subspecies strains; line 2 detects C. nebraskensis strains, and; line C is a control line. The primer/probe sets for genus Clavibacter and C. nebraskensis are labeled with Biotin/DIG and Biotin/FAM, respectively. A tenfold genomic DNA serial dilution (10 ng–10 fg) was used to perform the sensitivity assays. (A) Limit of detection determination using pure C. nebraskensis (A6094) DNA; detected until 100 fg and 10 pg using Clavibacter and C. nebraskensis specific primers/probe set, respectively. (B) Spiked multiplex test performed by adding 1 µL of corn plant sap in tenfold serially diluted C. nebraskensis (A6094) genomic DNA; detected until 1000 fg and 10 pg using Clavibacter and C. nebraskensis specific primers/probe set, respectively. (C) Detection using tenfold serially diluted C. michiganensis (A2058) purified genomic DNA; detected until 10 pg by Clavibacter genus-specific primers/probe—no detection with C. nebraskensis specific primers/probe set.