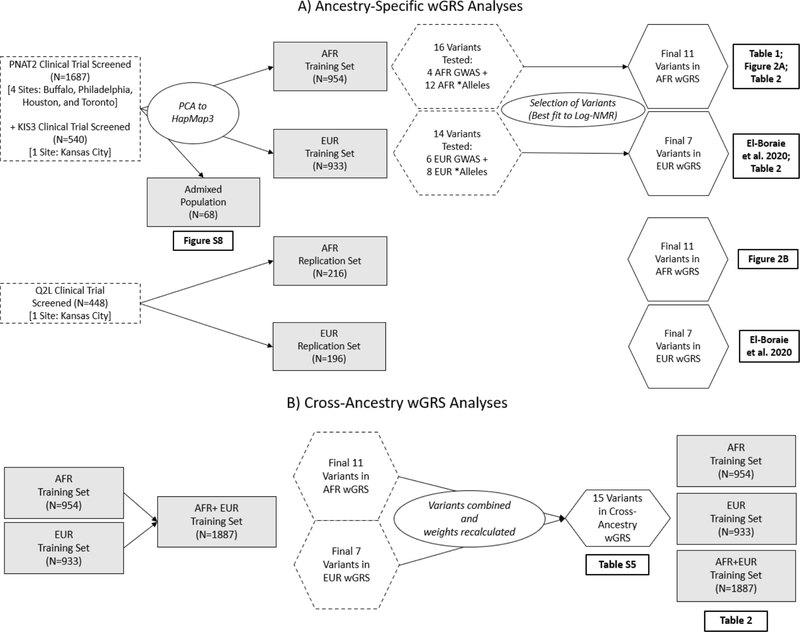
Figure 1.
Flowchart for the A) Ancestry-specific and B) Cross-ancestry wGRS analyses. The training sets consisted of individuals screened from two trials (PNAT2 and KIS3) and where ancestry was determined through PC clustering analysis to the HapMap3 populations. Variants tested included GWAS independent signals and * alleles from the specified population. For the cross-ancestry wGRS, all ancestry-specific wGRS variants were merged.