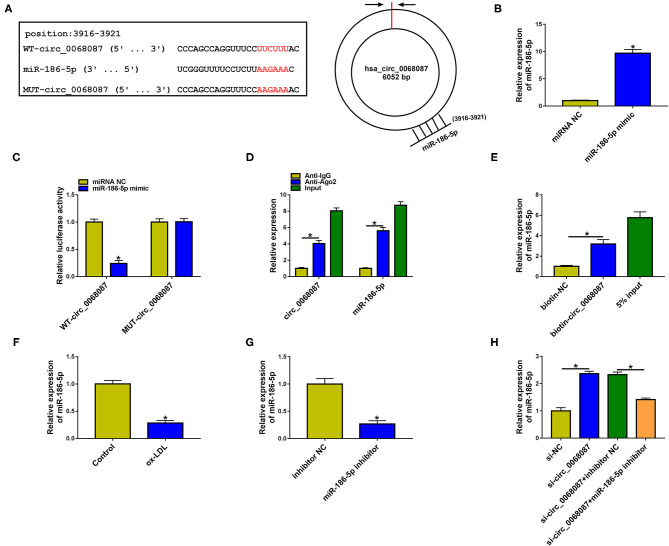
Figure 3.
Circ_0068087 acts as miR-186-5p sponge in HUVECs. (A) The putative binding sites between circ_0068087 and miR-186-5p were predicted by circinteractome and circBank. (B) The overexpression efficiency of miR-186-5p mimic in HUVECs was assessed by RT-qPCR. This experiment was performed three times with three technical repetitions each time. An unpaired Student t-test was used to evaluate the differences. (C,D) The predicted interaction between circ_0068087 and miR-186-5p was tested by dual-luciferase reporter assay and RIP assay. These two experiments were performed three times with three technical repetitions each time. An unpaired Student t-test was used to evaluate the differences. (E) RNA pulldown assay was applied to test the interaction between circ_0068087 and miR-186-5p. This experiment was performed three times with three technical repetitions each time. An unpaired Student t-test was used to evaluate the differences. (F) The level of miR-186-5p was measured in HUVECs exposed to 40 mg/L ox-LDL for 24 h by RT-qPCR. This experiment was performed three times with three technical repetitions each time. An unpaired Student t-test was used to evaluate the differences. (G) The silence efficiency of miR-186-5p inhibitor in HUVECs was assessed by RT-qPCR. This experiment was performed three times with three technical repetitions each time. An unpaired Student t-test was used to evaluate the differences. (H) The expression of miR-186-5p in HUVECs transfected with si-NC, si-circ_0068087, si-circ_0068087 + inhibitor NC, and si-circ_0068087 + miR-186-5p inhibitor was tested by RT-qPCR. This experiment was performed three times with three technical repetitions each time. One-way ANOVA followed by Tukey's post hoc test was used to assess the differences. *P < 0.05.